When FamilyTreeDNA released the new Mitotree, they also introduced their new mtDNA Discover tool, which is a series of 13 reports about each haplogroup, including one titled Ancient Connections.
Ancient Connections shows you ancient relatives from your direct matrilineal line through a mitochondrial DNA test or through a Y-DNA (preferably Big Y-700) test.
Ancient Connections help you connect the present to the past based on archaeological excavations around the world and DNA sequencing of remains. Ancient Connections links you through your DNA to ancient people, cultures, and civilizations that would be impossible to discover any other way. You don’t have to wonder if it’s accurate, or which line it came from, because you know based on the test you took. Discover’s Ancient Connections track the journey of your ancestors and relatives.
Ancient Connections can be very exciting – and it’s easy to get swept away on a wave of jubilation.
Are those people your ancestors, or relatives, or what? How do you know? How can you figure it out?
So let me just answer that question generally before we step through the examples, so you can unveil your own connections.
- You are RELATED to both Ancient and Notable Connections. Notable Connections are famous or infamous people who have lived more recently, and their relatives have been tested to identify their haplogroups.
- It’s VERY unlikely that Ancient Connections are your direct ancestors – but someone in the line that you share IS your ancestor.
- Many factors enter into the equation of how you are related, such as the haplogroup(s), the timeframe, and the location.
- The sheer number of people who were living at any specific time makes it very unlikely that any one person with that haplogroup actually was your direct ancestor. They are much more likely to be your distant cousin.
Factors such as whether you share the same haplogroup, similar locations, and the timeframe make a huge difference. Everyone’s situation is different with each Ancient Connection.
Ok, are you ready for some fun???
Let’s find out how to leverage these tools.
Ancient Connections
Ancient connections are fun and can also be quite useful for genealogy.
In this article, I’m going to use a mitochondrial DNA example because full sequence testers at FamilyTreeDNA just received their new Mitotree haplogroup. mtDNA Discover was released with Mitotree, so it’s new too. However, the evaluation process is exactly the same for Y-DNA.
Everyone’s results are unique, so your mileage absolutely WILL vary. What we are going to learn here is a step-by-step analytical process to make sure you’re hearing the message from your ancestors – and interpreting it correctly.
To learn about your new mitochondrial DNA haplogroup and haplotype, read the articles:
- New Mitotree Haplogroups and How to Utilize Them for Genealogy
- Mitochondrial DNA: What is a Haplotype Cluster and How Do I Find and Use Mine?
Radegonde Lambert
Let’s start with an Acadian woman by the name of Radegonde Lambert. She’s my ancestor, and I wrote about her years ago in the article, Radegonde Lambert (1621/1629-1686/1693), European, Not Native.
At the time, that article caused a bit of a kerfluffle, along with the article, Haplogroup X2b4 is European, Not Native American, because Radegonde’s X2b4 haplogroup had been interpreted by some to mean that her matrilineal ancestors were Native American.
That often happens when a genealogical line abruptly ends and hits a brick wall. What probably began with “I wonder if…”, eventually morphed into “she was Native,” when, in fact, she was not. In Radegonde’s case, it didn’t help any that her haplogroup was X2b4, and some branches of base haplogroup X2 are in fact Native, specifically X2a, However, all branches of X2 are NOT Native, and X2b, which includes X2b4, is not.
The Acadians were French people who established a colony in what is now Nova Scotia in the 1600s. They did sometimes intermarry with the Native people, so either Native or European heritage is always a possibility, and that is exactly why DNA testing is critically important. Let’s just say we’ve had more than one surprise.
I always reevaluate my own work when new data becomes available, so let’s look to see what’s happening with Radegonde Lambert now, with her new haplogroup and mtDNA Discover.
Sign on and Identify Your Haplogroup
You can follow along here, or sign on to your account at FamilyTreeDNA.
The first step is to take note of your new Mitotree haplogroup.
Your haplogroup badge is located near the bottom right of your page after signing in.
The tester who represents Radegonde Lambert has a Legacy Haplogroup of X2b4 and has been assigned a new Mitotree haplogroup of X2b4g.
Click Through to Discover
To view your personal Discover information, click on the Discover link on your dashboard.
You can simply enter a haplogroup in the free version of mtDNA Discover, but customers receive the same categories, but significantly more information if they sign in and click through.
You can follow along on the free version of Discover for haplogroups X2b4 here, and X2b4g here.
Clicking on either the Time Tree, or the Classic Tree shows that a LOT has changed with the Mitotree update.
Each tree has its purpose. Let’s look at the Classic Tree first.
The Classic Tree
I like the Classic Tree because it’s compact, detailed and concise, all in one. Radegonde Lambert’s new haplogroup, X2b4g is a subgroup of X2b4, so let’s start there.
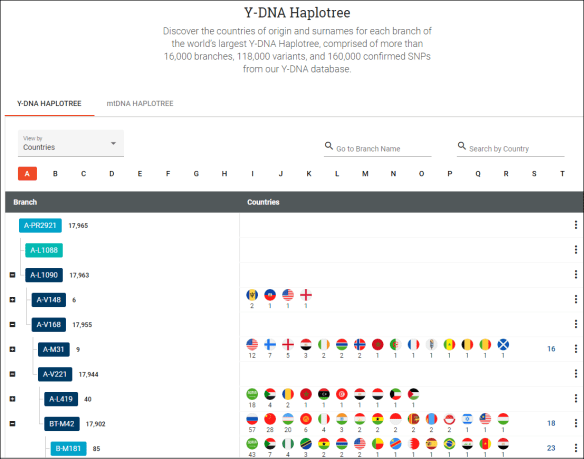
Under haplogroup X2b4, several countries are listed, including France. There are also 7 haplotype clusters, which tell you that those testers within the cluster all match each other exactly.
It’s worth noting that the little trowels (which I thought were shovels all along) indicate ancient samples obtained from archaeological digs. In the Discover tools, you’ll find them under Ancient Connections for that haplogroup. We will review those in a minute.
In Mitotree, haplogroup X2b4 has now branched several granular and more specific sub-haplogroups.
Radegonde Lambert’s new haplogroup falls below another new haplogroup, X2b4d’g, which means that haplogroup X2b4d’g is now the parent haplogroup of both haplogroups X2b4d and X2b4g. Both fall below X2b4d’g.
Haplogroup names that include an apostrophe mean it’s an umbrella group from which the two haplogroups descend – in this case, both X2b4d and X2b4g. Apostrophe haplogroups like X2b4d’g are sometimes referred to as Inner Haplogroups.
You can read more about how to understand your haplogroup name, here.
In this case, haplogroup X2b4d’g is defined by mutation G16145A, which is found in both haplogroups X2b4d and X2b4g. Both of those haplogroup have their own defining mutations in addition to G16145A, which caused two branches to form beneath X2b4d’g.
You can see that Radegonde Lambert’s haplogroup X2b4g is defined by mutation C16301T, but right now, that really doesn’t matter for what we’re trying to accomplish.
In descending order, for Radegonde, we have haplogroups:
- X2b4
- X2b4d’g
- X2b4g
Your Match Page
Looking at the tester’s match page, Radegonde’s haplotype cluster number and information about the cluster are found below the haplogroup. You can view your cluster number on:
- Your match page
- The Match Time Tree beside your name and those of your matches in the same haplotype cluster
- The Scientific Details – Variants page
I wrote about haplotype clusters, here.
On your match page, which is where most people look first, you are in the same haplogroup and haplotype cluster with anyone whose circle is also checked and is blue. If the little circles are not checked and blue, you don’t share either that haplogroup, haplotype cluster, or haplogroup and haplotype cluster. If you share a haplotype cluster, you will always share the same haplogroup.
Haplotype clusters are important because cluster members match on exactly the same (but less stable) mutations IN ADDITION to haplogroup-defining (more stable) mutations.
However, you may also share an identifiable ancestor with people in different haplotype clusters. Mutations, and back mutations happen – and a lot more often at some mutation locations, which is why they are considered less stable. Normally, though, your own haplotype cluster will hold your closest genealogical matches.
In Discover, you can see that Radegonde’s haplotype cluster, F585777, displays three tester-supplied countries, plus two more. Click on the little plus to expand the countries.
What you’re viewing are the Earliest Known Ancestor (EKA) countries that testers have entered for their direct matrilineal ancestor.
Let’s hope they understood the instructions, and their genealogy information was accurate.
Notice that Canada and France are both probably quite accurate for Radegonde, based on the known history of the Acadians. There were only French and Native women living in Nova Scotia in the 1600s, so Radegonde had to be one or the other.
The US may be accurate for a different tester whose earliest known ancestor (EKA) may have been found in, say, Louisiana. Perhaps that person has hit a brick wall in the US, and that’s all they know.
The US Native American flag is probably attributable to the old “Native” rumor about Radegonde, and the tester didn’t find the Canadian First Nations flag in the “Country of Origin” dropdown list. Perhaps that person has since realized that Radegonde was not Native and never thought to change their EKA designation.
The little globe with “Unknown Origins” is displayed when the tester doesn’t select anything in the “Country of Origin.”
Unfortunately, this person, who knew when Radegonde Lambert lived, did not complete any additional information, and checked the “I don’t know this information” box. Either Canada, or France would have been accurate under the circumstances. If they had tracked Radegonde back to Canada and read about her history, they knew she lived in Canada, was Acadian, and therefore French if she was not Native. Providing location information helps other testers, whose information, in turn, helps you.
Please check your EKA, and if you have learned something new, PLEASE UPDATE YOUR INFORMATION by clicking on the down arrow by your user name in the upper right hand corner, then Account Settings, then Genealogy, then Earliest Known Ancestors.
Don’t hesitate to email your matches and ask them to do the same. You may discover that you have information to share as well. Collaboration is key.
Radegonde’s Discover Haplogroup
First, let’s take a look at Radegonde’s haplogroup, X2b4g, in Discover.
The Discover Haplogroup Story landing page for haplogroup X2b4g provides a good overview. Please READ this page for your own haplogroup, including the little information boxes.
The history of Radegonde’s haplogroup, X2b4g, is her history as well. It’s not just a distant concept, but the history of a woman who is the ancestor of everyone in that haplogroup, but long before surnames. Haplogroups are the only way to lift and peer behind the veil of time to see who our ancestors were, where they lived, and the cultures they were a part of.
We can see that Radegonde’s haplogroup, X2b4g, was born in a woman who lived about 300 CE, Common (or Current) Era, meaning roughly the year 300, which is 1700 years ago, or 1300 years before Radegonde lived.
- This means that the tester shares a common ancestor with everyone, including any X2b4g remains, between now and the year 300 when haplogroup X2b4g was born.
- This means that everyone who shares haplogroup X2b4g has the same common female ancestor, in whom the mutation that defines haplogroup X2b4g originated. That woman, the common ancestor of everyone in haplogroup X2b4g, lived about the year 300, or 1700 years ago.
- Your common ancestor with any one individual in this haplogroup can have lived ANYTIME between very recently (like your Mom) and the date of your haplogroup formation.
- Many people misinterpret the haplogroup formation date to mean that’s the date of the MRCA, or most recent common ancestor, of any two people. It’s not, the haplogroup formation date is the date when everyone, all people, in the haplogroup shared ONE ancestor.
- The MRCA, or most recent common ancestor, is your closest ancestor in this line with any one person, and the TMRCA is the “time to most recent common ancestor.” It could be your mother, or if your matrilineal first cousin tested, your MRCA is your grandmother, and the TMRCA is when your grandmother was born – not hundreds or thousands of years ago.
- Don’t discount mitochondrial DNA testing by thinking that your common ancestor with your matches (MRCA) won’t be found before the haplogroup birth date – the year 300 in Radegonde’s case. The TMRCA for all of Radegonde’s descendants is about 1621 when she was born.
- The haplogroup birth date, 1700 years ago, is the common ancestor for EVERYONE in the haplogroup, taken together.
- Mitochondrial DNA is useful for BOTH recent genealogy and also reveals more distant ancestors.
- Looking back in time helps us understand where Radegonde’s ancestors lived, which cultures they were part of, and where.
There are two ways to achieve that: Radegonde’s upstream or parent haplogroups, and Ancient Connections.
Parent Haplogroups
X2b4g split from X2b4d’g, the parent haplogroup of BOTH X2b4d and X2b4g, around 3700 years ago, or about 1700 BCE (Before Common (or Current) Era).
Looking at either the Classic Tree, the Time Tree (above) or the Match Time Tree, you can see that haplogroup X2b4g has many testers, and none provide any locations other than France, Canada, the US, unknown, and one Native in the midst of a large haplotype cluster comprised of French and Canadian locations. Due to the size of the cluster, it’s only partially displayed in the screen capture above.
You can also see that sister haplogroup X2b4d split from X2b4d’g around the year 1000, and the ancestors of those two testers are reported in Norway.
Many, but not all of the X2b4g testers are descendants of Radegonde. Even if everyone is wrong and Radegonde is not French, that doesn’t explain the other matches, nor how X2b4g’s sister haplogroup is found in Norway.
Clearly, Radegonde isn’t Native, but there’s still more evidence to consider.
Let’s dig a little deeper using Radegonde’s Ancient Connections.
Ancient Connections
While ancestor and location information are user-provided, Ancient Connections are curated from scientifically published papers. There’s no question about where those remains were found.
When signed in to your account, if you’ve taken the mtFull Sequence test, clicking on the Ancient Connections tab in Discover shows a maximum of around 30 Ancient Connections. If you’re viewing the free version of Discover, or you’ve only tested at the HVR1 or HVR1+HVR2 levels, you’ll see two of your closer and one of your most distant Ancient Connections. It’s easy to upgrade to the mtFull.
In Discover, the first group of Ancient Connections are genetically closest to you in time, and the last connections will be your most distant. Some connections may be quite rare and are noted as such.
Please keep in mind that oldest, in this case, Denisova 8 and Sima de los Huesos, will never roll off your list. However, as new studies are released and the results are added to the tree, you may well receive new, closer matches. New results are being added with each Discover update.
It’s very exciting to see your Ancient Connections, but I need to say three things, loudly.
- Do NOT jump to conclusions.
- These remains are probably NOT YOUR ANCESTORS, but definitely ARE your distant cousins.
- Ancient Connections ARE wonderful hints, especially when taken together with each other and additional information.
It’s VERY easy to misinterpret Ancient Connections because you’re excited. I’ve done exactly that. To keep the assumption monster from rearing its ugly head, I have to take a breath and ask myself a specific set of questions. I step through the logical analysis process that I’m sharing with you.
The first thing I always want to know is where the genetically closest set of remains was found, when, and what we know about them, so let’s start there. Keep in mind that the closest remains genetically may not be the most recent set of remains to have lived. For example, my own haplogroup will be the closest genetically, but that person may have lived 2000 years ago. An Ancient Connection in a more distant haplogroup may have lived only 1000 years ago. The closest person genetically is NOT the same as the person who lived the most recently.
Our tester, Radegonde’s descendant, has no Ancient Connections in haplogroup X2b4g or X2b4d’g, but does have two in haplogroup X2b4, so let’s start there.
Discover provides a substantial amount of information about each set of ancient remains. Click on the results you want to view, and the information appears below.
Radegonde’s first Ancient Connection is Carrowkeel 534. The graphic shows the tester, the Ancient Connection being viewed, and their shared ancestor’s haplogroup. In this case, the shared ancestor haplogroup of Carrowkeel 534 and the tester is X2b4, who lived about 5000 years ago.
It’s very easy to look at Carrowkeel 534, become smitten, and assume that this person was your ancestor.

By Shane Finan – Own work, CC BY-SA 4.0, https://commons.wikimedia.org/w/index.php?curid=35098411
It’s especially easy if you WANT that person to be your ancestor. Carrowkeel 534 was buried in a passage tomb in County Sligo, Ireland. I’ve been there.
However, don’t let your emotions get involved – at least not yet.
This is the first example of the steps that determine that these remains are NOT YOUR ANCESTOR.
- Carrowkeel 534 was a male, and we all know that males do not pass on their mitochondrial DNA. Well, that’s an inconvenient fact.😊
- There are two sets of X2b4 remains in Ancient Connections. Carrowkeel 534 remains are about 4600-5000 years old, and your common ancestor with them lived about 5000 years ago. However, Radegonde was French and migration from Ireland to France is not typical.
- The other set of X2b4 remains, Ladoga 16, lived more recently, between the years of 900 and 1200 (or 800-1100 years ago), but they are found in Russia.
- Radegonde’s parent haplogroup, X2b4d’g was born about 3700 years ago, which excludes the Russian remains from being Radegonde’s direct ancestor.
- Radegonde’s common ancestor with both these sets of remains lived about 5000 years ago, but these remains were not found even close to each other.
In fact, these remains, if walking, are about 3299 km (2049 miles) apart, including two major water crossings.
- Given that Radegonde is probably French, finding her ancestor around 5000 years ago in an Irish passage tomb in County Sligo, or in a location east of St. Petersburg, is extremely unlikely.
What IS likely, though, is that X2b4d’g descendants of your common ancestor with both sets of remains, 5000 years ago, went in multiple directions, meaning:
- Radegonde’s ancestor found their way to France and along the way incurred the mutations that define X2b4d’g and X2b4g by the year 1600 when she lived, or about four hundred years ago.
- Another X2b4 descendant found their way to what is today Ireland between 4600 and 5000 years ago
- A third X2b4 descendant found their way to Russia between 800-1100 years ago, and 5000 years ago
If any question remains about the genesis of Radegonde’s ancestors being Native, Ancient Connections disproves it – BUT – there’s still an opportunity for misunderstanding, which we’ll see in a few minutes.
Ancient Connections Analysis Chart
I’ve created an analysis chart, so that I can explain the findings in a logical way.
Legend:
- Hap = Haplogroup
- M=male
- F=female
- U=unknown
Please note that ancient samples are often degraded and can be missing important mutations. In other words, the tree placement may be less specific for ancient samples. Every ancient sample is reviewed by FamilyTreeDNA’s genetic anthropologist before it’s placed on the tree.
Ancient samples use carbon dating to determine ages. Sometimes, the carbon date and the calculated haplogroup age are slightly “off.” The haplogroup age is a scientific calculation based on a genetic clock and is not based on either genealogy or ancient burials. The haplogroup age may change as the tree matures and more branches are discovered.
I’m dividing this chart into sections because I want to analyze the findings between groups.
The first entry is the earliest known ancestor of the current lineage – Radegonde Lambert, who was born about 1621, or roughly 400 years ago. I’ve translated all of the years into “years ago” to avoid any confusion.
If you wish to do the same, with CE (Current or Common Era) dates, subtract the date from 2000. 300 CE= (2000-300) or1700 years ago. With BCE dates, add 2000 to the BCE number. 1000 BCE= (1000+2000) or 3000 years ago.
| Connection Identity | Age Years Ago | Location & Cultural Group | Hap | Hap Age Years Ago | Shared Hap | Shared Hap Age Years Ago |
| Radegonde Lambert (F) | 400 | France or Canada -Acadian | X2b4g | 1700 | X2b4 | 5000 |
| Carrowkeel 534 (M) | 4600-5100 | Sligo, Ireland – Neolithic Europe | X2b4 | 5000 | X2b4 | 5000 |
| Ladoga 16 (M) | 800-1100 | Ladoga, Russia Fed – Viking Russia | X2b4 | 5000 | X2b4 | 5000 |
- Age Years Ago – When the Ancient Connection lived
- Hap Age Years Ago – When the haplogroup of the Ancient Connection (X2b4) originated, meaning was born
- Shared Hap Age Years Ago – When the Shared Ancestor of everyone in the Shared Haplogroup originated (was born)
In this first section, the haplogroup of the Ancient Connections and the Shared Haplogroup is the same, but that won’t be the case in the following sections. Radegonde Lambert’s haplogroup is different than her shared haplogroup with the Ancient Connections.
Let’s assume we are starting from scratch with Radegonde.
The first question we wanted to answer is whether or not Radegonde is European, presumably French like the rest of the Acadians, or if she was Native. That’s easy and quick.
Native people crossed Beringia, arriving from Asia someplace between 12,000 and 25,000 years ago in multiple waves of migration that spread throughout both North and South America.
Therefore, given that the first two samples, Carrowkeel 534 and Ladoga 16, share haplogroup X2b4, an upstream haplogroup with Radegonde Lambert, and haplogroup X2b4 was formed around 5000 years ago, the answer is that Radegonde’s X2b4 ancestor, whoever that was, clearly lived in Europe, NOT the Americas.
According to Discover, Haplogroup X2b4:
- Was formed about 5000 years ago
- Has 16 descendant haplogroups
- Has 29 unnamed lineages (haplotype clusters or individuals with no match)
- Includes testers whose ancestors are from 23 countries
The Country Frequency map shows the distribution of X2b4, including all descendant haplogroups. Please note that the percentages given are for X2b4 as a percentage of ALL haplogroups found in each colored country. Don’t be misled by the relative physical size of the US and Canada as compared to Europe.
The table view shows the total number of self-identified locations of the ancestors of people in haplogroup X2b4 and all downstream haplogroups.
The Classic Tree that we looked at earlier provides a quick view of X2b4, each descendant haplogroup and haplotype cluster, and every country provided by the 331 X2b4 testers.
For the X2b4 Ancient Connections, we’ve already determined:
- That Radegonde’s ancestors were not Native
- Carrowkeel 534 is a male and cannot be Radegonde’s ancestor. It’s extremely likely that Carrowkeel 534’s mother is not Radegonda’s ancestor either, based on several factors, including location.
- Based on dates of when Ladoga 16 lived, and because he’s a male, he cannot be the ancestor of Radegonde Lambert.
Radegonda’s haplogroup was formed long before Ladoga 16 lived. Each Ancient Connection has this comparative Time Tree if you scroll down below the text.
- Both Carrowkeel and Ladoga share an ancestor with our tester, and Radegonde, about 5000 years ago.
Think about how many descendants the X2b4 ancestor probably had over the next hundreds to thousands of years.
- We know one thing for sure, absolutely, positively – X2b4 testers and descendant haplogroups live in 32 countries. People migrate – and with them, their haplogroups.
What can we learn about the genealogy and history of Radegonde Lambert and her ancestors?
We find the same haplogroup in multiple populations or cultures, at different times and in multiple places. Country boundaries are political and fluid. What we are looking for are patterns, or sometimes, negative proof, which is often possible at the continental level.
X2b4, excluding downstream haplogroups, is found in the following locations:
- Bulgaria
- Canada (2)
- Czech Republic
- England (2)
- Finland (2)
- France (3)
- Germany (4)
- Portugal
- Scotland (2)
- Slovakia (2)
- Sweden (2)
- UK (2)
- Unknown (11)
- US (2)
Note that there are three people in France with haplogroup X2b4 but no more refined haplogroup.
Looking at X2b4’s downstream haplogroups with representation in France, we find:
- X2b4a (none)
- X2b4b (none)
- X2b4b1 (1)
- X2b4d’g (none)
- X2b4d (none)
- X2b4g (24) – many from Radegonde’s line
- X2b4e and subgroups (none)
- X2b4f (none)
- X2b4j and subgroups (none)
- X2b4k (none)
- X2b4l (1)
- X2b4m (none)
- X2b4n and subgroups (none)
- X2b4o (none)
- X2b4p (none)
- X2b4r (none)
- X2b4+16311 (none)
I was hoping that there would be an Ancient Connection for X2b4, X2b4d’g, or X2b4g someplace in or even near France – because that makes logical sense if Radegonde is from France.
All I can say is “not yet,” but new ancient sites are being excavated and papers are being released all the time.
Ok, so moving back in time, let’s see what else we can determine from the next set of Ancient Connections. Haplogroup X2b1”64 was formed about 5050 years ago.
| Connection Identity | Age Years Ago | Location & Cultural Group | Hap | Hap Age Years Ago | Shared Hap | Shared Hap Age Years Ago |
| Radegonde Lambert (F) | 400 | France or Canada | X2b4g | 1700 | ||
| Carrowkeel 534 (M) | 5100-4600 | Sligo, Ireland – Neolithic Europe | X2b4 | 5000 | X2b4 | 5000 |
| Ladoga 16 (M) | 800-1100 | Ladoga, Russia Fed – Viking Russia | X2b4 | 5000 | X2b4 | 5000 |
| Parknabinnia 186 (M) | 5516-5359 | Clare, Ireland – Neolithic Europe | X2b1”64 | 5516-5259 | X2b1”64 | Before 5050 years ago |
| Rössberga 2 (M) | 5339-5025 | Vastergotland, Sweden – Funnel Beaker | X2b1”64 | 5516-5259 | X2b1”64 | Before 5050 |
| Rössberga 29 (M) | 5366-5100 | Vastergotland, Sweden – Funnel Beaker and Early Plague | X2b1”64 | 5516-5259 | X2b1”64 | Before 5050 |
| Rössberga 38 (M) | 5340-5022 | Vastergotland, Sweden – Funnel Beaker | X2b1”64 | 5516-5259 | X2b1”64 | Before 5050 |
| Monte Sirai 797263 (U) | 2600-2400 | Monte Sirai, Italy (Sardinia) – Phoenicians | X2b35a1 | 3350 | X2b1”64 | 5050 |
| Bogovej 361 (F) | 1000-1100 | Lengeland, Denmark – Viking Denmark | X2b1”64 | 5516-5259 | X2b1”64 | 5050 |
| Ladoga 410 (M) | 800-1000 | Leningrad Oblast, Russia – Viking Russia | X2b1”64 | 5516-5259 | X2b1”64 | 5050 |
Our first group ended with haplogroup X2b4, and our second group consists of haplogroup X2b1”64, the parent haplogroup of X2b4. X2b1”64 is a significantly larger haplogroup with many downstream branches found throughout Europe, parts of western Asia, the Levant, India, and New Zealand (which probably reflects a colonial era settler). The Country Frequency Map and Table are found here.
X2b1”64 is just slightly older than X2b4, but it’s much more widespread, even though they were born about the same time. Keep in mind that haplogroup origination dates shift as the tree is developed.
- These seven individuals who share X2b1”64 as their haplogroup could be related to each other individually, meaning their MRCA, anytime between when they lived and when their haplogroup was formed.
- The entire group of individuals all share the same haplogroup, so they all descend from the one woman who formed X2b1”64 about 5050 years ago. She is the shared ancestor of everyone in the haplogroup.
One X2b4 and one X2b1”64 individual are found in the same archaeological site in Russia. Their common ancestor would have lived between the time they both lived, about 800 years ago, to about 5000 years ago. It’s also possible that one of the samples could be incomplete.
A second X2b1”64 Ancient Connection is found in the Court Tomb in County Clare, Ireland, not far from the Carrowkeel 534 X2b4 site.
However, Monte Sirai is fascinating, in part because it’s not found near any other site. Monte Sirai is found all the way across France, on an island in the Tyrrhenian Sea.
It may be located “across France” today, but we don’t know that the Phoenician Monte Sirai site is connected with the Irish sites. We can’t assume that the Irish individuals arrived as descendants of the Monte Sirai people, even though it would conveniently fit our narrative – crossing France. Of course, today’s path includes ferries, which didn’t exist then, so if that trip across France did happen, it could well have taken a completely different path. We simply don’t know and there are very few samples available.
Three Ancient Connections are found in the Rössberga site in Sweden and another in Denmark.
Adding all of the Ancient sites so far onto the map, it looks like we have two clusters, one in the northern latitudes, including Denmark, Sweden, and Russia, and one in Ireland with passage burials, plus one single Connection in Monte Sirai.
If I were to approximate a central location between all three, that might be someplace in Germany or maybe further east. But remember, this is 5000 years ago and our number of samples, as compared to the population living at the time is EXTREMELY LIMITED.
Let’s move on to the next group of Ancient Connections, who have different haplogroups but are all a subset of haplogroup X2.
| Identity | Age Years Ago | Location & Cultural Group | Hap | Hap Age Years Ago | Shared Hap | Shared Hap Age Years Ago |
| Radegonde Lambert (F) | 400 | France or Canada | X2b4g | 1700 | ||
| Carrowkeel 534 (M) | 5100-4600 | Sligo, Ireland – Neolithic Europe | X2b4 | 5000 | X2b4 | 5000 |
| Ladoga 16 (M) | 800-1100 | Ladoga, Russia Fed – Viking Russia | X2b4 | 5000 | X2b4 | 5000 |
| Parknabinnia 186 (M) | 5516-5359 | Clare, Ireland – Neolithic Europe | X2b1”64 | 5516-5259 | X2b1”64 | Before 5050 |
| Ross Rössberga 2 (M) | 5339-5025 | Vastergotland, Sweden – Funnel Beaker | X2b1”64 | 5516-5259 | X2b1”64 | Before 5050 |
| Rössberga 29 (M) | 5366-5100 | Vastergotland, Sweden – Funnel Beaker and Early Plague | X2b1”64 | 5516-5259 | X2b1”64 | Before 5050 |
| Rössberga 38 (M) | 5340-5022 | Vastergotland, Sweden – Funnel Beaker | X2b1”64 | 5516-5259 | X2b1”64 | Before 5050 |
| Monte Sirai 797263 (U) | 2600-2400 | Monte Sirai, Italy (Sardinia) – Phoenicians | X2b35a1 | 3350 | X2b1”64 | 5050 |
| Bogovej 361 (F) | 1000-1100 | Lengeland, Denmark – Viking Denmark | X2b1”64 | 5516-5259 | X2b1”64 | 5050 |
| Ladoga 410 (M) | 800-1000 | Leningrad Oblast, Russia – Viking Russia | X2b1”64 | 5516-5259 | X2b1”64 | 5050 |
| Barcin 31 (M) | 8236-8417 | Derekoy, Turkey – Neolithic Anatolia Ceramic | X2m2’5’7^ | 9200 | X2b”aq | 13,000 |
| Abasar 55 (M) | 500-800 | Abasár Bolt-tető, Abasar, Hungary – Medieval Hungary | X2m1e | 5350 | X2b”aq | 13,000 |
| Gerdrup 214 | 3779-3889 | Gerdrup, Sealand, Denmark – Middle Bronze Age | X2c1 | 3400 | X2+225 | 13,000 |
| Sweden Skara 275 | 800-1100 | Varnhem, Skara, Sweden – Viking Sweden | X2c1 | 3400 | X2+225 | 13,000 |
| Kopparsvik 225 | 950-1100 | Gotland, Sweden – Viking Sweden | X2z | 5650 | X2+225 | 13,000 |
| Sandomierz 494 | 900-1100 | Sandomierz, Poland – Viking Poland | X2c2b | 1650 | X2+225 | 13,000 |
| Kennewick man | 8390-9250 | Kennewick, Washington – Native American | X2a2’3’4^ | 10,450 | X2 | 13,000 |
| Roopkund 39 | 80-306 | Roopkund Lake, Uttarakhand, India – Historical India | X2d | 13,000 | X2 | 13,000 |
The next several Ancient Connections have haplogroups that are a subgroup of haplogroup X2. These people lived sometime between 500 years ago in Hungary, and 8390-9250 years ago when Kennewick Man lived in the present-day state of Washington in the US. Kennewick Man merits his own discussion, so let’s set him aside briefly while we discuss the others.
The important information to be gleaned here isn’t when these people lived, but when Radegonde shared a common ancestor with each of them. The shared haplogroup with all of these individuals was born about 13,000 years ago.
Looking at the map again, and omitting both X2 samples, we can see that the descendants of that shared ancestor 13,000 years ago are found more widely dispersed.
Including these additional burials on our map, it looks like we have a rather large Swedish and Viking cluster, where several of the older burials occurred prior to the Viking culture. We have a Southeastern Europe cluster, our two Irish tomb burials, and our remaining single Monte Sirai Phoenician burial on the island of Sardinia.
Stepping back one more haplogroup to X2, which was born about the same time, we add a burial in India, and Kennewick Man.
The Migration Map
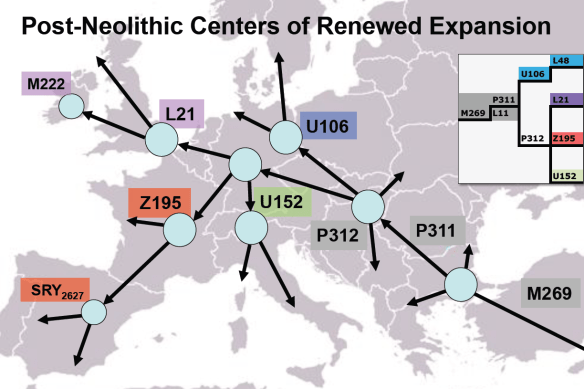
The Migration map in Discover provides two different features.
- The first is the literal migration map for the various ancestral haplogroups as they migrated out of Africa, if in fact yours did, culminating in your base haplogroup. In this case, the base haplogroup is X2, which is shown with the little red circle placed by FamilyTreeDNA. I’ve added the red squares, text and arrows for emphasis.
- The second feature is the mapped Ancient Connections, shown with little brown trowels. Clicking on each one opens a popup box.
After haplogroup X2 was formed, it split into haplogroups X2a and X2b.
The X2a group, Kennewick Man’s ancestors, made their way eastward, across eastern Russia to Beringia where they crossed into the Americas.
They either crossed Beringia, follow the Pacific coastline, or both, eventually making their way inland, probably along the Hood River, to where Kennewick Man was found some 2,800 years later on the banks of the Kennewick River.
The X2b group made their way westward, across western Europe to a location, probably France, where Radegonde Lamberts’ ancestors lived, and where Radegonde set sail for Nova Scotia.
After being separated for nearly 13,000 years, the descendants of the single woman who founded haplogroup X2 and lived someplace in central Asia around 13,000 years ago would find themselves on opposite coasts of the same continent.
So, no, Radegonde Lambert was not Native American, but her 600th matrilineal cousin or so, Kennewick Man, absolutely was.
Radegonde Lambert and Kennewick Man
Here’s where confirmation bias can rear its ugly head. If you’re just scanning the Ancient Connections and see Kennewick Man, it would be easy to jump to conclusions, leap for joy, slap a stamp of “confirmed Native American” on Radegonde Lambert, and never look further. And if one were to do that, they would be wrong.
Let’s work through our evaluation process using Discover.
Radegonde Lambert and Kinnewick Man, an early Native American man whose remains were found Kennewick, Washington in 1996, are both members of the broader haplogroup X2. Kennewick Man lived between 8290 and 9350 years ago, and their shared ancestor lived about 13,000 years ago – in Asia, where mitochondrial haplogroup X2 originated. This is the perfect example of one descendant line of a haplogroup, X2 in this case, going in one direction and a second one traveling in the opposite direction.
Two small groups of people were probably pursuing better hunting grounds, but I can’t help but think of a tundra version of the Hatfields and McCoys and cousin spats.
“I’m going this way. There are better fish on that side of the lake, and I won’t have to put up with you.”
“Fine, I’m going that way. There are more bears and better hunting up there anyway.”
Their wives, who are sisters, “Wait, when will I ever see my sister again?”
One went east and one went west.
X2a became Native American and X2b became European.
Looking back at our information about Kennewick Man, his haplogroup was born significantly before he lived.
He was born about 8390-9250 years ago, so let’s say 8820 years ago, and his haplogroup was born 10,500 years ago, so about 1680 years before he lived. That means there were many generations of women who carried that haplogroup before Kennewick Man.
Let’s Compare
Discover has a compare feature.
I want to Compare Radegonde Lambert’s haplogroup with Kennewick Man’s haplogroup X2a2’3’4^.
The Compare tool uses the haplogroup you are viewing, and you enter a second haplogroup to compare with the first.
The ancestral path to the shared ancestor, meaning their shared haplogroup, is given for each haplogroup entered. That’s X2 in this case. Then, from the shared haplogroup back in time to Mitochondrial Eve.
I prefer to view this information in table format, so I created a chart and rounded the haplogroup ages above X2.
| Hap Age – Years Ago | Radegonde’s Line | Shared Ancestors and Haplogroups | Kennewick’s Line | Hap Age – Years Ago | ||
| 143,000 | mt-Eve | |||||
| 130,000 | L1”7 | |||||
| 119,000 | L2”7 | |||||
| 99,000 | L2’3’4’6 | |||||
| 92,000 | L3’4’6 | |||||
| 73,500 | L3’4 | |||||
| 61,000 | L3 | |||||
| 53,000 | N | |||||
| 53,000 | N+8701 | |||||
| 25,000 | X | |||||
| 22,500 | X1’2’3’7’8 | |||||
| 13,000 | X2 – Asia | |||||
| 13,000 | X2+225 | X2a | 10,500 | |||
| 12,900 | X2b”aq | X2a2’3’4^ | 10,400 | Kennewick Man born c 8800 years ago | ||
| 11,000 | X2b | |||||
| 5,500 | X2b1”64 | |||||
| 5,000 | X2b4 | |||||
| 1,900 | X2b4d’g | |||||
| Radegonde Lambert born c 1661 – 400 years ago | 1,700 | X2b4g |
More Ancient Connections
Radegonde Lambert’s matrilineal descendants have an additional dozen Ancient Connections that are found in upstream haplogroup N-8701. Their shared ancestors with Radegonde reach back to 53,000 years ago in a world far different than the one we inhabit today. I’m not going to list or discuss them, except for one.
| Identity | Age Years Ago | Location & Cultural Group | Hap | Hap Age Years Ago | Shared Hap | Shared Hap Age Years Ago |
| Radegonde Lambert (F) | 400 | France or Canada | X2b4g | 1700 | ||
| Carrowkeel 534 (M) | 5100-4600 | Sligo, Ireland – Neolithic Europe | X2b4 | 5000 | X2b4 | 5000 |
| Ladoga 16 (M) | 800-1100 | Ladoga, Russia Fed – Viking Russia | X2b4 | 5000 | X2b4 | 5000 |
| Parknabinnia 186 (M) | 5516-5359 | Clare, Ireland – Neolithic Europe | X2b1”64 | 5516-5259 | X2b1”64 | Before 5050 |
| Rössberga 2 (M) | 5339-5025 | Vastergotland, Sweden – Funnel Beaker | X2b1”64 | 5516-5259 | X2b1”64 | Before 5050 |
| Rössberga 29 (M) | 5366-5100 | Vastergotland, Sweden – Funnel Beaker and Early Plague | X2b1”64 | 5516-5259 | X2b1”64 | Before 5050 |
| Rössberga 38 (M) | 5340-5022 | Vastergotland, Sweden – Funnel Beaker | X2b1”64 | 5516-5259 | X2b1”64 | Before 5050 |
| Monte Sirai 797263 (U) | 2600-2400 | Monte Sirai, Italy (Sardinia) – Phoenicians | X2b35a1 | 3350 | X2b1”64 | 5050 |
| Bogovej 361 (F) | 1000-1100 | Lengeland, Denmark – Viking Denmark | X2b1”64 | 5516-5259 | X2b1”64 | 5050 |
| Ladoga 410 (M) | 800-1000 | Leningrad Oblast, Russia – Viking Russia | X2b1”64 | 5516-5259 | X2b1”64 | 5050 |
| Barcin 31 (M) | 8236-8417 | Derekoy, Turkey – Neolithic Anatolia Ceramic | X2m2’5’7^ | 9200 | X2b”aq | 13,000 |
| Abasar 55 (M) | 500-800 | Abasár Bolt-tető, Abasar, Hungary – Medieval Hungary | X2m1e | 5350 | X2b”aq | 13,000 |
| Gerdrup 214 | 3779-3889 | Gerdrup, Sealand, Denmark – Middle Bronze Age | X2c1 | 3400 | X2+225 | 13,000 |
| Kopparsvik 225 | 950-1100 | Gotland, Sweden – Viking Sweden | X2z | 5650 | X2+225 | 13,000 |
| Sandomierz 494 | 900-1100 | Sandomierz, Poland – Viking Poland | X2c2b | 1650 | X2+225 | 13,000 |
| Sweden Skara 275 | 800-1100 | Varnhem, Skara, Sweden – Viking Sweden | X2c1 | 3400 | X2+225 | 13,000 |
| Kennewick man | 8390-9250 | Kennewick, Washington – Native American | X2a2’3’4^ | 10,450 | X2 | 13,000 |
| Roopkund 39 | 80-306 | Roopkund Lake, Uttarakhand, India – Historical India | X2d | 13,000 | X2 | 13,000 |
| Ranis 10 | 43,500-47,000 | Ranis, Germany – LRJ Hunger Gatherer | N3’10 | 53,000 | N+8701 | 53,000 |
| Zlatý kůň woman | 47,000 | Czech Republic – | N+8701 | 53,000 | N+8701 | 53,000 |
Zlatý kůň Woman
Zlatý kůň Woman lived some 43,000 years ago and her remains were discovered in the Czech Republic in 1950.
Believed to be the first anatomically modern human to be genetically sequenced, she carried about 3% Neanderthal DNA. Europeans, Asians and indigenous Americans carry Neanderthal DNA as well.
Unlike many early remains, Zlatý kůň Woman’s facial bones have been scanned and her face approximately reconstructed.
There’s something magical about viewing a likeness of a human that lived more than 40,000 years ago, and to whom I’m at least peripherally related.
Like all other Ancient Connections, it’s unlikely that I descend from Zlatý kůň Woman herself, but she is assuredly my very distant cousin.
What else do we know about Zlatý kůň Woman? Quoting from her Ancient Connection:
She lived during one of the coldest periods of the last ice age, surviving in harsh tundra conditions as part of a small hunter-gatherer group. She died as a young adult, though the cause of death remains unknown.
Her brain cavity was larger than that of modern humans in the comparative database, another trait showing Neanderthal affinity. While the exact colors of her features cannot be determined from available evidence, researchers created both a scientific grayscale model and a speculative version showing her with dark curly hair and brown eyes.
Zlatý kůň Woman may or may not have direct descendants today, but her haplogroup ancestors certainly do, and Radegonde Lambert is one of them, which means Radegonde’s matrilineal ancestors and descendants are too.
Ancient Connections for Genealogy
While Ancient Connections are fun, they are more than just amusing.
You are related through your direct matrilineal (mitochondrial) line to every one of your mtDNA Discover Ancient Connections. Everyone, males and females, can take a mitochondrial DNA test.
I find people to test for the mitochondrial DNA of each of my ancestral lines – like Radegonde Lambert, for example. I wrote about various methodologies to find your lineages, or people to test for them, in the article, Lineages Versus Ancestors – How to Find and Leverage Yours.
Radegonde’s mitochondrial DNA is the only key I have into her past, both recent and distant. It’s the only prayer I have of breaking through that brick wall, now or in the future.
Interpreted correctly, and with some luck, the closer Ancient Connections can provide genealogical insight into the origins of our ancestors. Not just one ancestor, but their entire lineage. While we will never know their names, we can learn about their cultural origins – whether they were Vikings, Phoenicians or perhaps early Irish buried in Passage Graves.
On a different line, an Ancient Connection burial with an exact haplogroup match was discovered beside the Roman road outside the European town where my ancestral line was believed to have been born.
Ancient Connections are one small glimpse into the pre-history of our genetic line. There are many pieces that are missing and will, in time, be filled in by ancient remains, Notable Connections, and present-day testers.
Check your matches and your Ancient Connections often. You never know when that magic piece of information you desperately need will appear.
What is waiting for you?
_____________________________________________________________
Share the Love!
You’re always welcome to forward articles or links to friends and share on social media.
If you haven’t already subscribed (it’s free,) you can receive an e-mail whenever I publish by clicking the “follow” button on the main blog page, here.
You Can Help Keep This Blog Free
I receive a small contribution when you click on some of the links to vendors in my articles. This does NOT increase your price but helps me keep the lights on and this informational blog free for everyone. Please click on the links in the articles or to the vendors below if you are purchasing products or DNA testing.
Thank you so much.
DNA Purchases and Free Uploads
- FamilyTreeDNA – Y, mitochondrial and autosomal DNA testing
- MyHeritage DNA – Autosomal DNA test
- MyHeritage FREE DNA file upload – Upload your DNA file from other vendors free
- AncestryDNA – Autosomal DNA test
- AncestryDNA Plus Traits
- 23andMe Ancestry – Autosomal DNA only, no Health
- 23andMe Ancestry Plus Health
Genealogy Products and Services
- MyHeritage Subscription with Free Trial
- Legacy Family Tree Webinars – Genealogy and DNA classes, subscription-based, some free
- Legacy Family Tree Software – Genealogy software for your computer
- OldNews – Old Newspapers with links to save to MyHeritage trees
- MyHeritage Omni comprehensive “everything included” subscription plan
- Newspapers.com – Search newspapers for your ancestors
- NewspaperArchive – Search different newspapers for your ancestors
My Books
- DNA for Native American Genealogy – by Roberta Estes, for those ordering the e-book from anyplace, or paperback within the United States
- DNA for Native American Genealogy – for those ordering the paperback outside the US
- The Complete Guide to FamilyTreeDNA – Y-DNA, Mitochondrial, Autosomal and X-DNA – for those ordering the e-book from anyplace, or paperback within the United States
- The Complete Guide to FamilyTreeDNA – Y-DNA, Mitochondrial, Autosomal and X-DNA for those ordering the paperback from outside the US
Genealogy Books
- Genealogical.com – Lots of wonderful genealogy research books
- American Ancestors – Wonderful selection of genealogy books
Genealogy Research
- Legacy Tree Genealogists – Professional genealogy research