Sometimes we’ve been doing genetic genealogy for so long we forget what it’s like to be new. I’m reminded, sometimes humorously, by some of the questions I receive.
When I do DNA Reports for clients, each person receives a form to complete with a few questions designed to give them the opportunity to tell me what their testing goals are and to ask any questions they might have. One woman asked, “Can you tell me about my psychogroup?”
I thought that psychogroup was particularly appropriate for a cluster of genealogists, especially genetic genealogists, but decided I had better let that one go.
Then there was the person who wanted to know about their hologroup. I wondered if I needed 3D glasses for that one.
Someone else wondered about their helpgroup. I couldn’t help but think of introducing myself, “Hello, my name is Roberta and I’m a member of haplogroup J.” Kinda gives new meaning to “what’s your sign?”
Then there was the person who though it was a Biblical reference, Holygroup and wanted to know how they connected to Biblical folks. Well, we do talk about Y-line Adam and Mitochondrial Eve, so why wouldn’t someone ask that?
My favorite, though is the person who gave this reason for leaving a haplogroup project, “not my glopo.” Hey, at least they realized that, as opposed to the person who called me a member of the KKK for suggesting that they did not belong in a particular project. I found that to be particularly humorous, given my ethnic mix, heritage and family.
But today, when my cousin asked me if a haplogroup follows the mitochondrial DNA, I decided it was time to talk about what a haplogroup is, a little history, and why we use them. And Shanen, thanks for asking!
Think of a haplogroup as an ancestral clan, a large family, like the Celts, or Vikings. These would be larger than Native American tribes, encompassing members of many tribes. There are two male Native American haplogroups that include all Native American males. There are a few more African clans, or haplogroups, but not many.
There are clans for the Y chromosome, which is of course tested by the Y DNA test at Family Tree DNA and generally follows the paternal surname up and down the tree. Y DNA is passed from father to son, only, through the Y chromosome which only males possess.
There are also clans for mitochondrial DNA, tested by the mtDNA tests at Family Tree DNA, which follows the direct maternal line up your family tree. This means your mother, her mother, her mother, etc. Woman give their mitochondrial DNA to all of their offspring, males and females, but only females pass it on.
You can see the Y-line, paternal (blue) and mitochondrial, maternal (red), lines on the pedigree chart below.
Companies like 23andMe and the Geno 2.0 project provide haplogroups for both Y-line and mitochondrial DNA, but neither of them test personal mutations that allow you to compare your mutations against those of other people for genealogical matches. The regular Y-line and mitochondrial tests at Family Tree DNA do that. In addition, both also provide your haplogroup or clan designation.
A new haplogroup is born when a very specific new mutation occurs. All descendants will carry that mutation. That mutation defines that haplogroup. So if a new haplogroup is born today, we wouldn’t know it was a haplogroup until hundreds or thousands of years later when we see that lots of people have this same mutation from a single individual. As you might imagine, many haplogroups over the ages have died out, but some have been very successful as evidenced by the fact that we are all here today! Roughly half of the European men carry Y haplogroup R and mitochondrial haplogroup H is found in nearly 50% of all Europeans – both descending, respectively, from one single person tens of thousands of years ago.
Since all of humanity, both male and female, sprang initially from Africa, the earliest haplogroups were found there. As some people moved further away and crossed into Asia and Europe, they developed unique mutations that would give rise to the European, Asian and Native American haplogroups we know today. There are 4 main groupings, African, European, Asian and Native American, but there are several subgroups within most of those main groups, except for Native Americans who only have two male haplogroups.
So in essence, haplogroups are a pedigree chart of the clans of humanity. Family Tree DNA displays a haplogroup chart with the main haplogroups shown on everyone’s personal page for Y-line DNA. They were simply named alphabetically with no connection to a word. So no, A is not A because it’s African, even though it happens to be. N is not Native American. E is not European. You get the drift. Any resemblance is purely coincidental.
Your clan, in this example, haplogroup I, is shown with an arrow. Every clan, male and female, has subclans, often known as subclades for Y DNA or subgroups for mitochondrial DNA. To see the various subgroups of I, click on the tab and voila, there they are, the subgroups of haplogroup I. Yours is the lowest one on the tree that is green, in this case I2b1a1.
Because of the dramatic new number of haplogroups recently discovered, future versions of the haplotree will be moving away from the letter based names like I2b1a1 and will only use the terminal, or lowest branch, SNP to identify a haplogroup. In this case, that would be L126 or L137 which are equivalent SNPs. So in the future this person’s haplogroup will be called I-L126 instead of I2b1a1 because L126 will never change, but the name I2b1a1 changes every time a new upstream haplogroup is discovered between the root of haplogroup I and I2b1a1 and needs to be inserted into its proper place in the tree.
As we learn more about the subgroups, each one has its own story which is somewhat different than the stories of the other subgroups. Some are evident, such as Jewish clusters, some not so much. Each clan story involves how that haplogroup came to be found where it is today. For example, haplogroup E is African, but within haplogroup E, there are two major divisions with very different stories for their clans. One group is found only in Sub-Saharan African and one is found mostly in the Middle East and the Mediterranean basin and is known colloquially as the Berber haplogroup. We’re still learning about subgroups, and with the Geno 2.0 test, the haplotree is growing exponentially.
Family Tree DNA predicts your clan, or haplogroup, with any Y-DNA test as long as you match exactly at 12 markers to someone who has been SNP tested. SNP testing is what tests for the special haplogroup defining mutations. If you don’t match, they will SNP test you for free to establish your primary haplogroup.
Many people purchase additional SNP tests to further define their Y haplogroup so that they can learn about where their ancestors were, when, and what they were doing. For example, we know that SNP M222 equates to Niall of the 9 Hostages in history. How cool is that to know!
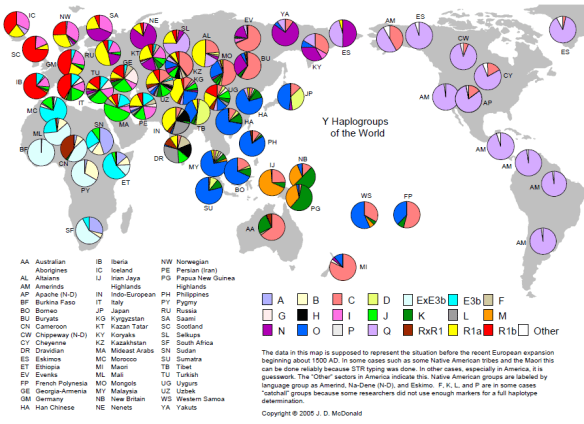
Some years ago, Dr. Doug McDonald assembled this wonderful map of the basic haplogroups of the world. Although we’ve discovered subgroups for each haplogroup, it’s still quite valid. E3b has since been renamed E1b1b and ExE3b means haplogroup E1b1a. RxR1 means haplogroup R except R1a and R1b which have their own legend.
Mitochondrial DNA also has haplogroups, which are clans. On the drawing below, compliments of Dr. Whit Athey, it’s easier to see how the daughter clans arose, were born, and were named. Because of the naming pattern, this looks a little less like a pedigree chart and a little more like stars, planets and moons.
One difference between Y-line DNA and mitochondrial DNA clans is that although they are all currently named alphabetically, the mitochondrial clans have names. That is thanks to Dr. Bryan Sykes who wrote the book, “The Seven Daughters of Eve” published in 2001. For example, he named haplogroup H, Helena because Helena is Greek for light. He told somewhat accurate stories about each clan and although quite scientifically dated now, described the life that each clan would have lived in post-glacial Europe. This book was the first book about DNA to reach the popular reading public, and was a huge success because he humanized science and normal air-breathing humans could relate. I ordered my first mitochondrial DNA test through his company and received one page with a Sunday School gold star on the red dot for haplogroup J.
I was thrilled at the time, but times have changed a lot. Due to advances in research and new subclades being defined, thanks in large part to citizen scientists like you, I now know that I’m haplogroup J1c2f as a result of my full sequence mitochondrial DNA test.
Unlike Y-line DNA, no additional SNP test is required to fully determine your mitochondrial DNA haplogroup. When you take the full mitochondrial sequence test (mtFullSequence) at Family Tree DNA, you receive your most detailed, full haplogroup designation automatically. With the HVR1 (mtDNA) and HVR2 (mtDNAPlus) tests, you receive at least your base haplogroup. The full sequence is required to determine your full haplogroup.
To put this in perspective, think of your mitochondrial DNA as a clock face. There are a total of 16,569 locations in your mitochondrial DNA. The HVR1 test tests the number of locations from 11:55 to noon and the HVR2 test tests the number of locations between noon and 12:05PM. The full sequence test tests the rest, the balance of the 50 minutes of the hour.
Family Tree DNA is the only commercial testing company that offers the full sequence test.
As more discoveries are made for both male and female haplogroups, the subgroup names sometimes change within the clan or main haplogroup because new branches get inserted in the tree as they are discovered.
For example, from a scientific paper, here’s an early version of the haplogroup H mitochondrial phylotree which is what the haplotree is called for mitochondrial DNA.
Here’s a later version.
You wouldn’t even be able to see today’s version, because the print would have to be miniscule to fit it on the page. In Dr. Behar’s paper, “A ‘Copernican’ Reassessment of the Human Mitochondrial DNA Tree from its Root” published in April 2012, the supplemental material records haplogroup H87. Most of those subgroups have subgroups of their own, like you can see above, and those that don’t today soon will as new discoveries are made.
Now that you know what a haplogroup is, there’s a lot you can do with both mitochondrial and Y-line DNA results.
Even if you do nothing more, it’s fun to identify your clan. It’s the only way of extending our genealogy back in time beyond surnames. For me, to connect my last known maternal ancestor, Elisabetha Mehlheimer, born in or near Goppsmannbuhl, Germany around 1800 to the cave paintings in Chauvet, France created about 12,000 years ago was a magical moment, a reach across time through a tenuous umbilical strand allowing me to identify and touch my 12,000 year-old ancestor. In my wildest genealogist dreams, I never dreamed this could or would ever be possible and indeed, it wouldn’t be, were it not for the genetic genealogy tools we have today.
- ______________________________________________________________
Disclosure
I receive a small contribution when you click on some of the links to vendors in my articles. This does NOT increase the price you pay but helps me to keep the lights on and this informational blog free for everyone. Please click on the links in the articles or to the vendors below if you are purchasing products or DNA testing.
Thank you so much.
DNA Purchases and Free Transfers
- Family Tree DNA
- MyHeritage DNA only
- MyHeritage DNA plus Health
- MyHeritage FREE DNA file upload
- AncestryDNA
- 23andMe Ancestry
- 23andMe Ancestry Plus Health
- LivingDNA
Genealogy Services
Genealogy Research
- Legacy Tree Genealogists for genealogy research