Y-DNA Haplogroup O has been found in male testers descended from a Native American ancestor, or in Native American tribes in the Americas – but sometimes things are more complex than they seem. The story of when and how haplogroup O arrived in the Americas is fascinating – and not at all what you might think.
Introduction
The concept of Native American heritage and indigenous people can be confusing. For example, European Y-DNA haplogroup R is found among some Native American men. Those men may be tribal members based on their mother’s line, or their haplogroup R European Y-DNA may have been introduced either through adoption practices or traders after the arrival of Europeans.
There is unquestionable genetic evidence that the origin of Haplogroup R in the Americas was through colonization, with no evidence of pre-contact indigenous origins.
Y-DNA testing and matching, specifically the Big Y-700 test, with its ability to date the formation of haplogroups very granularly, has successfully identified the genesis of Y-DNA haplogroups and their movement through time.
We’ve spent years trying to unravel several instances of Native American Y-DNA Haplogroup O and their origins. Native American, in this context, means that men with haplogroup O are confirmed to be Native American at some point in documented records. This could include early records, such as court or probate records, or present-day members of tribes. There is no question that these men are recognized as Native American in post-contact records or are tribal members, or their descendants.
What has not been clear is how and when haplogroup O entered the Native American population of these various lineages, groups, or tribes. In other words, are they indigenous? Were they here from the earliest times, before the arrival of colonists, similar to Y-DNA haplogroups C and Q?
This topic has been of great interest for several years, and we have been waiting for additional information to elucidate the matter, which could manifest in several ways:
- Ancient pre-contact DNA samples of haplogroup O in the Americas, but none have been found.
- Current haplogroup O testers in Native American peoples across the North and South American continents, forming a connecting trail genetically, geographically, and linearly through time. This has not occurred.
- Big-Y DNA matches within the Americas between Haplogroup O Native American lines unrelated in a genealogical timeframe whose haplogroup formation pre-dates European contact. This has not occurred.
- Big-Y DNA matches between Haplogroup O men whose haplogroups were formed in the Americas after the Beringian migration and expansion that scientists agree occurred at least 12-16K years ago, and possibly began earlier. Earlier human lineages, if they existed, may not have survived. A later Inuit and Na-Dené speaker circumpolar migration occurred 4-7K years ago. This has not occurred.
- Big-Y DNA matches with men whose most recent common ancestor haplogroup formation dates connect them with continental populations in other locations, outside of North and South America. This would preclude their presence in the Americas after the migrations that populated the Americas. This has occurred.
The Beringian migration took place across a now-submerged land bridge connecting the Chutkin Peninsula in Russia across the Bering Strait with the Seward Peninsula in Alaska.

By Erika Tamm et al – Tamm E, Kivisild T, Reidla M, Metspalu M, Smith DG, et al. (2007) Beringian Standstill and Spread of Native American Founders. PLoS ONE 2(9): e829. doi:10.1371/journal.pone.0000829. Also available from PubMed Central., CC BY 2.5, https://commons.wikimedia.org/w/index.php?curid=16975303
Haplogroup O is clearly Native American in some instances, meaning that it occurs in men who are members of or descend from specific Native American tribes or peoples. One man, James Revels, is confirmed in court records as early as 1656. However, ancestors of James Revels fall into category #5, as their upstream parental haplogroup is found in the Pacific islands outside the Americas after the migration period.
Based on available evidence, the introduction of haplogroup O appears to be post-contact. Therefore, haplogroup O is not indigenous to the Americans in the same sense as haplogroups Q and C that are found widespread throughout the Americas in current testers who are tribal members, descendants of tribal members, and pre-contact ancient DNA as mapped in the book, DNA for Native American Genealogy.
Ancient DNA

Haplogroup C is found in both North and South America today, as are these ancient DNA locations.

Haplogroup Q is more prevalent than Haplogroup C, and ancient DNA remains are found throughout North and South America before colonization.
No ancient DNA for Haplogroup O has been discovered in the Americas. We do find contemporary haplogroup O testers in regional clusters, which we will analyze individually.
Let’s take a look at what we have learned recently.
Wesley Revels’ Lineage
Wesley Revels was the initial Y-DNA tester whose results identified Haplogroup O as Native American, proven by a court record. That documentation was critical, and we are very grateful to Wesley for sharing both his information and results.
Wesley’s ancestor, James Revels, was Native American, born about 1656 and bound to European planter, Edward Revell. James was proven in court to be an Accomack “Indian boy” from “Matomkin,” age 11 in 1667. James was bound, not enslaved, until age 24, at which time he was to be freed and receive corn and clothes.
James had died by 1681 when he was named several times in the Accomack County records as both “James, an Indian” and “James Revell, Indian,” in reference to his estate. James lived near Edward Revell, his greatest creditor and, therefore, administrator of his estate, and interacted with other Indian people near Great Matompkin Neck. Marie Rundquist did an excellent job of documenting that here. Additional information about the Revels family and Matomkin region can be found here.

The location where Edward Revell lived, Manokin Hundred, was on the water directly adjacent the Great Matomkin (now Folly Creek) and Little Matomkin Creeks, inside the Metomkin Inlet. The very early date tells us that James Revels’s paternal ancestor was in the colonies by 1656 and probably born about 1636, or perhaps earlier.
Lewis and Revels men are later associated with the Lumbee Tribe, now found in Robeson and neighboring counties in North Carolina. The Lewis line descends from the Revels lineage, as documented by Marie and Wesley. Other men from this line have tested and match on lower-level STR markers, but have not taken the much more granular and informative Big-Y test.
Until recently, the men who matched Wesley Revels closely on the Big-Y test were connected with the Revels line and/or the Lumbee.
Wesley has a 37-marker STR match to a man with a different surname who had not tested beyond that level, in addition to several 12-marker STR matches to men from various locations. Men who provided known ancestral or current locations include one from Bahrain, two from the Philippines, and three from China. Those men have not taken the Big-Y, and their haplogroups are all predicted from STR results to O-M175 which was formed in Asia about 31,000 years ago.
12-marker matches can reach thousands of years back in time. Unless the matches share ancestors and match at higher levels, 12-marker matches are only useful for geographic history, if that. The Big Y-700 test refines haplogroup results and ages from 10s of thousands of years to (generally) within a genealogically relevant timeframe, often within a couple hundred years.
One of Wesley’s STR matches, Mr. Luo, has taken a Big Y-700 test. Mr. Luo descends directly from Indonesia in the current generation and is haplogroup O-CTS716, originating about 244 BCE, or 2244-ish years ago. Mr. Luo does not match Wesley on the Big-Y test, meaning that Wesley and Mr. Luo have 30 or more SNP differences in their Big-Y results, which equates to about 1,500 years. The common ancestor of Wesley Revels and Mr. Luo existed more than 1,500 years ago in Indonesia. It’s evident that Mr. Luo is not Native American, but his location is relevant in a broader analysis.
There is no question that Wesley’s ancestor, James Revels, was Native American based on the court evidence. There is also no question that the Revels’ paternal lineage was not in the Americas with the Native American migration group 12-16K years ago.
The remaining question is how and when James Revels’ haplogroup O ancestor came to be found on the Atlantic seaboard in the early/mid 1600s, only a few years after the founding of Jamestown.
The results of other Haplogroup O men may help answer this question.
Mr. Lynn
Another haplogroup O man, Mr. Lynn, matches Wesley on STR markers, but not on the Big-Y test.
Mr. Lynn identified his Y-DNA line as Native American, although he did not post detailed genealogy. More specifically, we don’t know if Mr. Lynn identified that he was Native on his paternal line because he matches Wesley, or if the Native history information was passed down within his family, or from genealogical research. Mr. Lynn could also have meant generally that he was Native, or that he was Native “on Dad’s side,” not specifically his direct patrilineal Y-line.
Based on Mr. Lynn’s stated Earliest Known Ancestor (EKA) and additional genealogical research performed, his ancestor was John Wesley Lynn (born approximately 1861, died 1945), whose father was Victor Lynn. John’s death certificate, census, and his family photos on Ancestry indicate that he was African American. According to his death certificate, his father, Victor Lynn, was born in Chatham Co., NC, just west of Durham.

Family members are found in Baldwin Township, shown above.
I did not locate the family in either the 1860 or 1870 census. In 1860, the only Lynn/Linn family in Chatham County was 50-year-old Mary Linn and 17-year-old Jane, living with her, presumably a daughter. Both are listed as “mulatto” (historical term) with the occupation of “domestic.” They may or may not be related to John Wesley Lynn.
In 1870, the only Linn/Lynn in Chatham County is John, black, age 12 or 13 (so born in 1857 or 1858), farm labor, living with a white family. This is probably not John Wesley Lynn given that he is found with his mother in 1880 and the ages don’t match.
In 1880. I find Mary Lynn in Chatham County, age 48, single, black, with daughter Eliza Anne, 20, mulatto, sons John Wesley, 14 so born about 1866, and Charles 12, both black. Additionally, she is living with her nieces and nephews, Cephus, black, 12, Lizzie, 7, mulatto, Malcom, 4, mulatto, William H, 3, mulatto (I think, written over,) and John age 4, mulatto. The children aged 12 and above are farm labor.
In 1880, I also find Jack Lynn, age 28, black, married with 3 children, living beside William Lynn, 25, also married, but with no children.
Trying to find the family in 1870 by using first name searches only, I find no black Mary in 1870 or a mulatto Mary with a child named Jack or any person named Cephus by any surname. I don’t find Jack or any Lynn/Linn family in Chatham County.
The 1890 census does not exist.
In the 1900 census, I find Wesley Lynn in Chatham County, born in January of 1863, age 37, single, a boarder working on the farm of John Harris who lives beside Jack Lynn, age 43, born in April of 1857. Both Lynn men are black. I would assume some connection, given their ages, possibly or probably brothers.
In 1940, John Wesley Lynn, age 74, negro (historical term), is living beside Victor Lynn, age 37, most likely his son.
I could not find Victor Lynn, John Wesley Lynn’s father in any census, so he was likely deceased before 1880 but after 1867, given that Mary’s son Charles Lynn was born in 1868, assuming Mary’s children had the same father. The fact that Mary was listed as single, not married nor widowed suggests enslavement, given that enslaved people were prohibited from legally marrying.
About the only other assumption we can make about Victor Sr. is that he was probably born about 1832 or earlier, probably in Chatham County, NC based on John Wesley’s death certificate, and he was likely enslaved.
Subclades of Haplogroup O
Both the Revels and Lynn men are subclades of haplogroup O and both claim Native heritage – Wesley based on the Revels genealogy and court documents, and Mr. Lynn based on the Native category he selected to represent his earliest known paternal ancestor at FamilyTreeDNA.
Both men have joined various projects, including the American Indian Project, which provides Marie and me, along with our other project co-administrators, the ability to work with and view both of their results at the level they have selected.
How Closely Related Are These Haplogroup O Men?
How closely related are these two men?
- Do the haplogroups of the Revels men and Mr. Lynn converge in a common ancestor in a timeframe BEFORE colonialization, meaning before Columbus “discovered” the Caribbean islands when colonization and the slave trade both began?
- Do the haplogroups converge on North or South American soil or elsewhere?
- Is there anything in the haplogroup and Time Tree information that precludes haplogroup O from being Native prior to the era of colonization?
- Is there anything that confirms that a haplogroup O male or males were among the groups of indigenous people that settled the Americas sometime between 12 and 26 thousand years ago? Or even a later panArctic or circumpolar migration wave?
Haplogroup O is well known in East Asia, Indonesia, and the South Pacific.
Another potential source of haplogroup O is via Madagascar and the slave trade.

The Malagasy Roots Project has several haplogroup O individuals, including the Lynn and Revels men, who may have joined to see if they have matches. We don’t know why the various haplogroup O men in the project joined. Other haplogroup O men in the project may or may not have proven Malagasay heritage.
Information provided by the project administrators is as follows:
The people of Madagascar have a fascinating history embedded in their DNA. 17 known slave ships came from Madagascar to North America during the Transatlantic Slave Trade. As a result, we find Malagasy DNA in the African American descendants of enslaved people, often of Southeast Asian origin. One of the goals of this project is to discover the Malagasy roots of African Americans and connect them with their cousins from Madagascar. Please join us in this fascinating endeavor. mtDNA Haplogroups of interest include: B4a1a1b – the “Malagasy Motif”, M23, M7c3c, F3b1, R9 and others Y-DNA Haplogroups include: O1a2 – M50, O2a1 – M95/M88, O3a2c – P164 and others
Resources:
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2987306/ http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1199379/ http://mbe.oxfordjournals.org/cgi/pmidlookup?view=long&pmid=19535740 http://www.biomedcentral.com/1471-2156/15/77 http://www.biomedcentral.com/1471-2164/10/605
The Malagasy group only has one other man who is haplogroup O and took the Big-Y test, producing haplogroup O-FTC77008. Of course, we don’t know if he has confirmed Madagascar ancestry, and his haplogroup is quite distant from both Revels and Lynn in terms of when his haplogroup was formed.
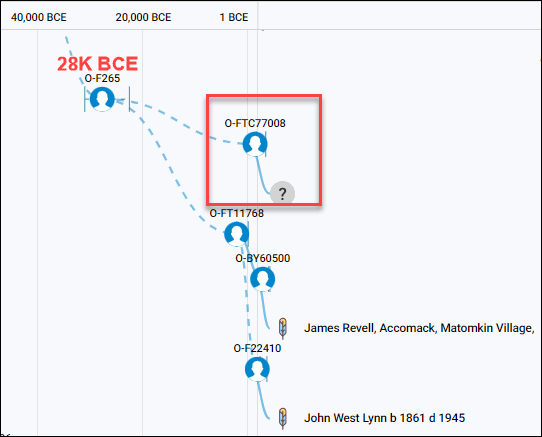
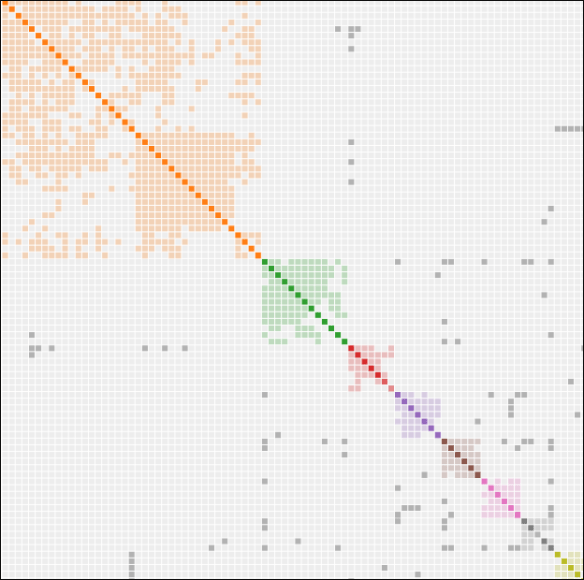
Viewing the Malagasy Project’s Group Time Tree, above, the common ancestor between those three men lived about 28K BCE, or 30,000 years ago.
Haplogroup O Project Group Time Tree
The Haplogroup O Project Time Tree provides a better representation of haplogroup O in general given that it has a much wider range of samples.

On this tree, I’ve labeled the haplogroup formation dates, along with the Revels/Lewis line which descends from O-FT45548. This haplogroup includes one additional group member whose surname is locked, as he hasn’t given publication permission. The haplogroup formation date of 1766 occurs approximately 85 years after James Revel’s birth, so is attributable to some, but not all of his descendants. At least one descendant falls into the older Haplogroup O-BY60500.
The common ancestor of all three, meaning Revels, Lewis, and the man whose name is locked and does not know his genealogy, is haplogroup O-BY60500, born about 1741.
Their ancestral haplogroup before that, O-FT11768, is much older.
Two Filipino results are shown on and descending from the parent branch of O-FT11768, formed about 3183 BCE, or about 5183 years ago. This tells us that the ancestors of all these men were in the same place, most likely the Philippines, at that time.
3183 BCE (5180 years ago) is well after the Native American migration into the Americas.
Discover Time Tree
Obviously, not every tester joins a project, so now I’m switching to the Discover Time Tree which includes all Y-DNA haplogroup branches. Their common haplogroup, O-FT11768, has many branches, not all of which are shown below. I’m summarized unseen branch locations at bottom left.

Expanding the Time Tree further to view all of the descendant haplogroups of O-FT11768, we see that this was a major branch with many South Pacific results, including the branch of O-FT22410, bracketed in red, which has three members.
One is Mr. Lynn whose feather indicates Native American as his EKA country selection, one is a man whose ancestor is from Singapore, and one is an unknown individual who did not enter his ancestor’s country of origin.
Geography

Wesley’s STR match list, which can reflect matches further back in time than the Big-Y test, includes islands near Singapore. This geography aligns with what is known about haplogroup O.
The distance between this Asian region and continental America, 9000+ miles distant by air, is remarkable and clearly only navigable at that time by ship, meaning ships with experienced crew, able to navigate long distances with supplies and water.
We know that in 760 CE, about 1240 years ago, Mr. Lynn’s haplogroup O-F24410 was formed someplace in the South Pacific – probably in Malaysia or a nearby island. This region, including the Philippines, is home to many haplogroup O men. The majority of haplogroup O is found in Asia, the South Pacific, and Diaspora regions.

We know that Hawaii was populated by Polynesian people about 1600 years ago, prior to the age of colonization. Hawaii is almost 7000 miles from Singapore.
Here’s the challenge. How did these haplogroup O men get from the South Pacific to Virginia? Mr. Lynn and the Singapore tester share a common ancestor about 1240 years ago, or 760 CE.
There is no known or theorized Native American settlement wave across Beringia as late as 760 CE. We know that the parent haplogroup was someplace near Singapore in approximately 760 CE.

Two Filipino men and the Revels’ ancestors were in the same location in the Pacific Islands 5180 years ago. How did they arrive on the Eastern Shore in Virginia, found in the Native population, either in or before 1656 when James Revels was born?
What happened in the 3500 years between those dates that might explain how James Revel’s ancestor made that journey?
Academic Papers
In recent years, there has been discussion of possible shoreline migration routes along the Russian coast, Island hopping along Alaska, Canada, and what is now the US, known as the Kelp Highway or Coastal Migration Route – but that has yet to be proven.
Even if that is the case, and it’s certainly a possibility, how did this particular group of men get from the Pacific across the continent to the Atlantic shore in such a short time, leaving no telltale signs along the way? The Coastal Migration Theory hypothesis states that this migration occurred from 12-16 thousand years ago, and then expanded inland over the next 3-5K years. They could not have expanded eastward until the glaciers receded. Regardless, the parent haplogroup and associated ancestors are still found in the Philippines and South Pacific 5000 years ago – after that migration and expansion had already occurred.
The conclusion of the paper is that there is no strong evidence for a Pacific shoreline migration. Regardless, that’s still thousands of years before the time range we’re observing.
We know that the Lynn ancestor was with men from Indonesia in 760 CE, and the Revels ancestor was with men from the Pacific Islands, probably the Philippines, 5180 years ago. They couldn’t have been in two places at the same time, so the ancestors of Revels and Lynn were not in the Americas then.
A 2020 paper shows that remains from Easter Island (Rapa Nui) show Native American DNA, and suggests that initial contact occurred between the two cultures about 1200 CE, or about 800 years ago, but there is not yet any pre-contact or post-contact ancient Y-DNA found in the Americas that shows Polynesian DNA. Furthermore, the hypothesis is that the DNA found on Easter Island came from the Americas, not vice versa. The jury is still out, but this does show that trans-Pacific contact between the two cultures was taking place 800 years ago, at least two hundred years pre-European contact.
Australasian migration to South America is also suggested by one set of remains found in Brazil dating from more than 9000 years ago, but there have been no other remains found indicating this heritage, either in Brazil, or elsewhere in the Americas.
Based on the Time Tree dates of the Haplogroup O testers in our samples, we know they were in the Islands of Southeast Asia after this time period. Additionally, there are no Australia/New Zealand matches.
The Spanish
The Spanish established an early trade route between Manila and Acapulco beginning in 1565. Consequently, east Asian men left their genetic signature in Mexico, as described in this paper.
Historians estimate that 40-129K immigrants arrived from Manilla to colonial Mexico between 1565 and 1815, with most being enslaved upon arrival. Approximately one-third of the population in Manilla was already enslaved. Unfortunately, this paper focused only on autosomal genome-wide results and did not include either Y-DNA, nor mitochondrial. However, the paper quantifies the high degree of trade, and indicates that the Philippines and other Asian population haplotypes are still prevalent in the Mexican population.
In 2016, Dr. Miguel Vilar, the lead scientist with the National Geographic Genographic project lectured in Guam about the surprising Native American DNA found in the Guam population and nearby islands. He kindly provided this link to an article about the event.

Guam was colonized by Spain. In the image from the Boxer Codex, above, the local Chamorro people greet the Manila Galleon in the Ladrones Islands, as the Marianas were called by the Spanish, about 1590.
Native Hawaiians descend from Polynesian ancestors who arrived in the islands about 400 CE, or about 1600 years ago. Captain Cook, began the age of European contact in Hawaii in 1778.
Five Possibilities
There are five possible origins of haplogroup O in the Americas.
- Traditional migration across Beringia with the known migrations, estimated to have occurred about 12-16K years ago.
- A Kelp Highway Coastal Migration which may have occurred about 12-16K years ago and dispersed over the next 3-5K years.
- Circumpolar migration – specifically Inuit and Na-Dene speakers, about 4-6K years ago.
- Post-contact incorporation from the Pacific Islands resulting from shipping trade on colonial era ships sometime after 1565.
- Post-contact incorporation from Madagascar resulting from the importation of humans who may or may not have been enslaved upon arrival.
Do we have any additional evidence?
Other Haplogroup O DNA
From my book, DNA for Native American Genealogy:
Testers in haplogroup O-BY60500 and subclade O-FT45548 have proven Native American heritage.
We have multiple confirmed men from a common ancestor who is proven to be an enslaved Accomack “Indian boy,” James Revell, born in 1656, “belonging to the Motomkin” village, according to the Accomack County, Virginia court records. These men tested as members of haplogroup O-F3288 initially, after taking the Big Y-500 test. However, upgrading to the Big Y-700 produced more granular results and branches reflecting mutations that occurred since their progenitor was born in 1656.
Unfortunately, other than known descendants, these men have few close Y-DNA or Big Y-700 matches.
Without additional men testing from different unrelated lines, or ancient haplogroup O being discovered, we cannot confirm that this haplogroup O male’s ancestor was not introduced into the Matomkin Tribe in some way post-contact. Today, one descendant from this line is a member of the Lumbee Tribe.
However, that isn’t the end of the haplogroup O story.
The Genographic Project data shows one Haplogroup O Tlingit tribal member from Taku, Alaska, along with several testers from Mexico that indicate their paternal line is indigenous. Some people from Texas identify their paternal line as Hispanic.
Another individual indicates they were born on the Fountain Indian Reserve, in British Columbia and speaks the St’at’imcets language, an interior branch of Coastal Salish.

Haplogroup O has been identified as Native American in other locations as well.
Much of the information about Haplogroup O testers was courtesy of the Genographic Project, meaning we can’t contact those people to request upgraded tests, and we can’t obtain additional information in addition to what they provided when they tested. As an affiliate researcher, I’m very grateful to the National Geographic Society’s Genographic project for providing collaborative data.
When the book was published, the Discover Time Tree had not yet been released. We have additional information available today, including the dates of haplogroup formation.
FamilyTreeDNA Haplotree and Discover
The FamilyTreeDNA Haplotree (not to be confused with the Discover Time Tree) shows 10 people at the O-M175 level in Mexico, 10 people in the US report Native American heritage, 2 in Jamaica, and one each in Peru, Panama, and Cuba. There’s also one tester from Madagascar.
Altogether, this gives us about 35 haplogroup O males in the Americas, several with Native heritage.
Please note that I’ve omitted Hawaii in this analysis and included only North and South America. The one individual selecting Native Hawaiian (Kanaka Maoli) is in haplogroup O-M133.
Let’s look at our three distinct clusters.
Cluster 1 – Pacific Northwest – Alaska and Canada

We have a cluster of three individuals along the Pacific Coast in Alaska and Canada who have self-identified as Native, provided a tribal affiliation, and, in some cases, the spoken language.
How might haplogroup O have arrived in or near Vancouver, Washington? We know that James Cook “discovered” Hawaii in 1778, naming it the Sandwich Islands. By 1787, a female Hawaiian died en route to the Pacific Northwest, and the following year, a male arrived. Hawaii had become a provisioning stop, and the Spanish took Hawaiians onto ships as replacement workers.
Hawaiian seamen, whalers, and laborers began intermarrying with the Native people along the West Coast as early as 1811. Their presence expanded from Oregon to Alaska. Migration and intermarriage along the Pacific coast began slowly, but turned into a steady stream 30 years later when we have confirmed recruitment and migration of Hawaiian people
In 1839, John Sutter recruited a small group of 10 Hawaiians to travel with him to the then-Mexican colony of Alta, California.
By the mid-1800s, hundreds of Hawaiians lived in Canada and California. In 1847, it was reported that 10% of San Francisco’s residents were Hawaiian. Some of those people integrated with the Native American people, particularly the Miwok and Maidu. The village of Verona, California was tri-lingual: Hawaiian, a Native language, and English, and is today the Sacramento-Verona Tribe.
This article provides a history of the British Company who administered Fort Vancouver, near Vancouver, Washington, that included French-Canadians, Native Americans and Hawaiians. In 1845, 119 Hawaiians were employed at the fort. One of the 119, Opunuia, had signed on as an “engagé,” meaning some type of hired hand or employee, with the Hudson Bay Company for three years, after which he would be free to return home to Honolulu or establish himself in the Oregon Country. He married a woman from the Cascade Tribe.
The descendants of the Hawaiian men and Native women were considered tribal members. In most tribes, children took the tribal status and affiliation of the mother.
The Taku and Sitka, Alaska men on the map are Tlingit, and the man from British Columbia is from the Fountain Indian Reserve.
Hawaiian recruitment is the most likely scenario by which haplogroup O arrived in the tribes of the Pacific Northwest. In that sense, haplogroup O is indeed Native American but not indigenous to that region. The origins of haplogorup O in the Pacific Northwest are likely found in Hawaii, where it is indigenous, and before that, Polynesia – not due to a Beringian crossing.
Cluster 2 – Mexico

We find a particularly interesting small cluster of 4 haplogroup O individuals in interior Mexico.
In the 1500s, Spain established a trade route between Mexico and Manilla in the Philippines.
In 1564, four ships left Mexico to cross the Pacific to claim Guam and the Philippines for King Philip II of Spain. The spice trade, back and forth between Mexico and the Philippines began the following year and continued for the next 250.
Landings occurred along the California coast and the western Mexican coastline. The majority of the galleon crews were Malaysian and Filipino who were paid less than the Spanish sailors. Slaves, including people from the Marianas were part of the lucrative cargo.

One individual in Texas reports haplogroup O and indicates their paternal ancestors were Hispanic/Native from Mexico. A haplogroup O cluster claiming Native heritage is found near Zacatecas, Fresnillo and San Luis Potosi in central Mexico. Additionally, mitochondrial haplogroup F, also Asian, is found there as well. Acapulco is the lime green pin.

An additional haplogroup O tester with Native heritage is found in Lima, Peru.
Haplogroup O men are found in Panama, Jamaica and Cuba, but do not indicate the heritage of their paternal ancestral line. None of these men have taken Big-Y tests, and some may well have arrived on the slave ships from Madagascar, especially in the Caribbean. This source attributes some enslaved people in Jamaica to Hawaiian voyages.
I strongly suspect that the Mexican/Peru grouping in close proximity to the Pacific coastline is the result of the Manilla-Mexico 250-year trade route. The Spanish also plied those waters regularly. Big Y testing of those men would help flesh-out their stories – when and how haplogroup O arrived in the local population.
Cluster 3 – East Coast
At first glance, the East Coast grouping of men with a genetic affinity to the people of the Philippines and Indonesia seems more difficult to explain, but perhaps not.
On the East Coast, we have confirmed reports of whalers near Nantucket as early as 1765 utilizing crewmen from Hawaii, known then as the Sandwich Islands, Tahiti, and the Cape Verde Islands off of Africa. A thorough review of early literature might well reveal additional information about early connections with the Sandwich Islands, and in particular, sailors, crew, or enslaved people.
The Spanish and French were the first to colonize the Philippines by the late 1500s. They had discovered the Solomon Islands, Melanesia, and other Polynesian Islands, and by the early 1600s, the Dutch were involved as well.
The Encyclopedia Britanica further reports that Vasco Balboa first sailed into the Pacific in 1513 and seven years later, Ferdinand Magellan rounded the tip of South America. The Spanish followed, establishing a galley trade between Manila, in the Philippines and Acapulco in western Mexico.
While I found nothing specific stating that the earliest voyages brought men from the Philippines and Oceania back to their European home ports with them, we know that early European captains on exploratory voyages took Native people from the east coast of the Americas on their return journey, so there’s nothing to preclude them from doing the same from the Pacific. The early explorers stayed for months among the Oceanic Native peoples. If they were short on sailors for their return voyage, Polynesian men filled the void.
We know that the Spanish took slaves as part of their trade. We know that the ships in the Pacific took sailors from the islands. If the men themselves didn’t stay in the locations they visited, it’s certainly within the realm of possibility that they fathered children with local, Native women. Furthermore, given that the slave trade was lucrative, it’s also possible that some Pacific Island slaves were taken not as crew but with the intention of being sold into bondage. Other men may have escaped the ships and hidden among the Native Tribes along the eastern seaboard.
Fishing in Newfoundland and exploration in what would become the US was occurring by 1500, so it’s certainly possible that some of the indigenous people from Indonesia and the Philippines were either stranded, sold to enslavers, escaped, or chose to join the Native people along the coastline in North America. Ships had to stop to resupply rations and take on fresh water.
We know that by the mid-1600s, James Revels, whose father carried haplogroup O, had been born on the Atlantic coast of Virginia or Maryland, probably on the Delmarva Peninsula, short for Delaware, Maryland, Virginia, where the Accomac people lived.
There are other instances of haplogroup O found along the east coast.

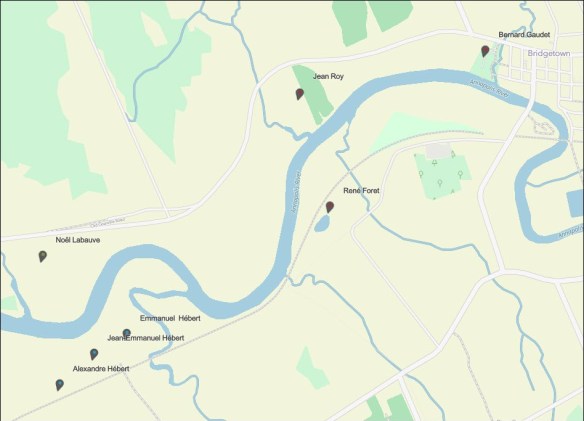
On the eastern portion of the haplogroup O map from the book, DNA for Native American Genealogy, we find the following locations:
- Hillburn, NY – man identified as “Native American Black.”
- Chichester County, PA – Genographic tester identified the location of his earliest known ancestor – included here because O is not typically found in the states.
- Accomack County, VA – Delmarva peninsula – James Revels lineage
- Robeson County, NC – Lewis and Revels surname associated with the Lumbee
- Chatham County, NC – Lynn ancestor’s earliest known location
- Greene County, NC – enslaved Blount ancestor’s EKA in 1849
The genesis of Mr. Blount’s enslaved ancestor is unclear. Fortunately, he took a Big Y-700 test.

Mr. Blount’s only Big-Y match is to a man from the United Arab Emirates (UAE), but the haplogroup history includes Thailand, which is the likely source of both his and his UAE matches’ ancestors at some point in time. Their common ancestor was in Thailand in 336 CE, almost 1700 years ago.

All surrounding branches of haplogroup O on the Time Tree have Asian testers, except for the one UAE gentleman.
The Blount Haplogroup O-FTC77008 does not connect with the common ancestral haplogroup of Lynn and Revels, so these lineages are only related someplace in Oceana prior to O-F265, or more about 30,000 years ago. Their only commonality other than their Asian origins is that they arrived on the East Coast of the Americas.
We know that the Spanish were exploring the Atlantic coastline in the 1500s and were attempting to establish colonies. In 1566, a Spanish expedition reached the Delmarva Peninsula. This spit of land was contested and changed hands several times, belonging variously to the Spanish, Dutch, and British by 1664.
Furthermore, we also know that the ships were utilizing slave labor. One of the Spanish ships wrecked in the waters off North Carolina near Hatteras or Roanoke Island before the Lost Colony was abandoned on Roanoke Island in 1587. The Croatan Indians reported that in memorable history, several men, some of whom were reported to be slaves, had survived the wreck and “disappeared” into the hinterlands – clearly running for their lives.
These men, if they survived, would have been incorporated into the Native population as there were no other settlements at the time. Variations of this scenario may have played out many times.
James Revels’ ancestor could have arrived on any ship, beginning with exploration and colonization in the early 1500s through the mid-1650s.
By the time the chief bound the Indian boy who was given the English name James to Edward Revell, James’s Oceanic paternal ancestor could have been 4, 5 or 6 generations in the past – or could have been his father.
The Accomack was a small tribe, loosely affiliated with the Powhatan Confederacy along the Eastern Shore. By 1700, their population had declined by approximately 90% due to disease. A subgroup, the Gingaskins, intermarried with African Americans living nearby. After Nat Turner’s slave rebellion of 1831, they were expelled from their homelands.
The swamps near Lumberton in Robeson County, NC, became a safe haven for many mixed-race Native, African, and European people. The swamps protected them, and they existed, more or less undisturbed, for decades. Revels and Lewis descendants are both found there.
Many Native Americans were permanently enslaved alongside African people – and within a generation or so, their descendants knew they were Native and African, but lost track of which ancestors descended from which groups. Life was extremely difficult back then. Generations were short, and enslaved people were moved from place to place and sold indiscriminately, severing their family ties entirely, including heritage stories.
Returning to the Discover Time Tree Maps
Wesley Revels has STR matches with several men from Indonesia, China, and the Philippines. It would be very helpful if those men would upgrade to the Big Y-700 so that we can more fully complete the haplogroup O branches of the Time Tree.

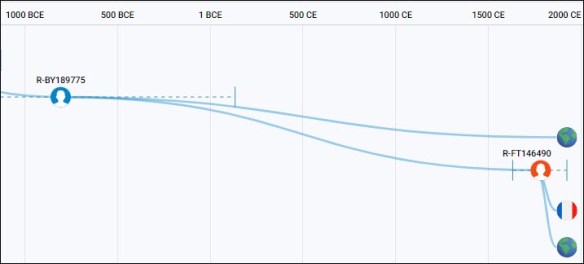
The common Revels/Lewis ancestor, accompanied by two descendant men on different genetic branches from the Philippines, was born about 5180 years ago. There is no evidence to suggest Haplogroup O-FT11768 was born anyplace other than in the Philippines.
How did the descendant haplogroups of O-FT45548 (Revels, Lewis, and an unnamed man) and O-F22410 (Lynn) arrive in Virginia or anyplace along the Atlantic seaboard?
Hawaii wasn’t settled until about 1600 years ago. We know Hawaiians integrated with the Pacific Coast Native tribes in the 1800s, but James Revels was in Virginia in 1656..
We know that the Spanish established a mid-1500s trade route between Manila and Acapulco, leaving their genetic signature in western Mexico.
None of these events fit the narrative for the Revels or the Lynn paternal ancestor.
Furthermore, the Revels and Lynn lines do not connect on North American soil, as both descend from the same parent haplogroup, O-FT11768, 5180 years ago in the Philippines. This location and history suggest a connection with the Spanish galleon trade era. The haplogroup formation clearly predates that trade, which means those men were still in the Philippines, not already living on the American continents. Therefore, the descendants of the haplogroup O-FT11768 arrived in Virginia and North Carolina sometime after that haplogroup formation 5100 years ago.

The Lynn ancestor connects with a man from Singapore in 760 CE, or just 1240 years ago. A descendant of haplogroup O-F22410 arrived in North Carolina sometime later.
It does not appear, at least not on the surface, that there is a connection through Madagascar, although we can’t rule that out without additional testers. If the connection is through Madagascar, then their ancestors were likely transported from Indonesia to Madagascar, then as enslaved people from Madagascar to the Atlantic colonies to be sold. However, James Revels was not enslaved. He was clearly Native and bound to a European plantation owner, who did, in fact, free him as agreed and subsequently loaned him money.
Based on the dates involved, and when we know they were in Oceania, an arrival along the west coast, followed by a quick migration across the country to a peninsula of land in the Atlantic, is probably the least likely scenario. There is also no historical or ancient haplogroup O DNA found anyplace between the west and east coasts, nor in the Inuit or Na-Dene speakers. The Navajo, who speak the Na-Dené language, migrated to the Southwest US around 1400 CE, but haplogroup O has not been found among Na-Dené speakers.
It’s a long way from Singapore and the Philippines to Madagascar, so while the coastal migration scenario is not impossible, it’s also not probable, especially given what we know about the Spanish Pacific trade that existed profitably for 250 years.
However, one haplogroup O subgroup arrived in the UAE by some methodology after 336 CE.
It’s entirely possible, indeed probable, that haplogroup O arrived in the Americas for various reasons, on different paths, in different timeframes.
Haplogroup O was found in people in the Americas after colonization had begun. There has been no ancient Haplogroup O DNA discovered, and there’s evidence indicating that these instances of haplogroup O could not have arrived in any of the known Beringia migrations nor the theorized Coastal or Kelp migration. We know the East Coast Cluster is not a result of the West Coast 19th-century migration because James Revels was in court one hundred and fifty years before the Hawaiians were living among the Native people along the Pacific coastline.
There’s nothing to indicate that the Mexican group that likely arrived beginning in the mid-1500s for the next 250 years as a result of the Indonesian trade route migrated to the east coast, or vice versa. That’s also highly unlikely.
The most likely scenario is that Mr. Lynn’s, Mr. Blount’s, and James Revels’ ancestors were brought on trade ships, either as sailors or enslaved men. They may not have stayed, simply visited. They may each have arrived in a completely different scenario, meaning Mr. Blount’s ancestors could have been enslaved arrivals from Madagascar, Mr. Lynn’s from Indonesia, and Mr. Revel’s as a crew member on a Spanish ship. We simply don’t know.
James Revels’ descendants were Native through his mother’s tribe, as confirmed in the 1667 court records. However, the Revels and Lynn lineages weren’t Native as a result of their paternal haplogroup O ancestors crossing Beringia into the Americas with Native American haplogroups Q and C. Instead, the Lynn and Revels migration story is quite different. Their ancestors arrived by ship. The journey was long, perilous, and far more unique than we could have imagined, taking them halfway around the world by water.
Timeline
There’s a lot of information to digest, so I’ve compiled a timeline incorporating both genetic and historical information for easy reference.
- 30,000 years ago (28,000 BCE) – haplogroup O-F265, common Asian ancestor of Mr. Blount, the Revels/Lewis group, Mr. Lynn, and an unknown Big-Y tester in the Malagasy group project
- 12,000-16,000 years ago – Indigenous Americans arrived across now-submerged Beringia
- 12,000-16,000 years ago – possible Coastal Migration route may have facilitated a secondary source of indigenous arrival along the Pacific coastline of the Americas
- 4000-7000 years ago – circumpolar migration arrival of Inuit and Na-Dené speakers found in the Arctic polar region and the Navajo in the Southwest who migrated from Alaska/Canada about 1400 CE
- 5180 years ago (3180 BCE) – haplogroup O-FT11768, the common ancestor of Mr. Lynn and the Revels/Lewis group with many subgroups in the Philippines, Hawaii, Singapore, Brunei, China, Sumatra, and Thailand
- 2244 years ago (244 BCE) – haplogroup O-CTS716, the common ancestor of Wesley Revels and Mr. Luo from Indonesia
- The year 336 CE, 1684 years ago – haplogroup O-FTC77008, the common ancestor of Mr. Blount, UAE tester and a man from Thailand
- 400 CE, 1600 years ago – Hawaii populated by Polynesian people
- 760 CE, 1240 years ago – haplogroup O-F22410, common ancestor of Mr. Lynn with a Singapore man
- 1492 CE, 528 years ago – Columbus begins his voyages to the “New World,” arriving in the Caribbean
- By 1504 CE – European fishing began off of Newfoundland
- 1565 – Spain claimed Guam and the Philippines
- 1565 – Spanish trade between Manilla and Acapulco begins and continues for 250 years, until 1815, using crews of men from Guam, the Philippines, and enslaved people from the Marianas.
- 1565 – St. Augustine (Florida) was founded by the Spanish as a base for trade and conquest along the eastern seaboard
- 1566 – A Spanish expedition reached the Delmarva peninsula intending to establish a colony, but bad weather thwarted that attempt.
- 1585-1587 – voyages of discovery by the English and the Lost Colony on Roanoke Island, North Carolina
- 1603 – English first explored the Delmarva Peninsula, home to the Accomac people, now Accomack County, VA, where James Revels’s court record was found in 1667
- 1607 – Jamestown, Virginia, founded by the English
- 1608 – Colonists first arrived on the Delmarva Peninsula and allied with Debedeavon, whom they called the “laughing King” of the Accomac people. At that time, the Accomac had 80 warriors. Debedeavon was a close friend to the colonists and saved them from a massacre in 1622. He died in 1657.
- 1620 – The Mayflower arrived near present-day Provincetown, Massachusetts
- 1631-1638 – Dutch West India Company established a colony on the Delmarva Peninsula, but after conflicts, it was destroyed by Native Americans in 1638. The Swede’s colony followed, and the region was under Dutch and Swedish control until it shifted to British control in 1664
- 1656 – Birth of James Revels, confirmed in a 1667 court record stating that he was an Accomack “Indian boy” from “Matomkin,” judged to be age 11, bound to Edward Revell. This location is on the Delmarva Peninsula.
- 1741 CE – Haplogroup O-BY60500 formation date that includes all of the Revels and Lewis testers who descend from James Revels born in 1656
- 1765 – Whalers near Nantucket using crewmen from Hawaii (Sandwich Islands), Tahiti, and the Cape Verde Islands off of Africa
- 1766 CE – Formation date for haplogroup O-FT45548, child haplogroup of O-BY60500, for some of the Lewis and Revels men who all descend from James Revels born in 1656
- 1778 – Captain Cook makes contact with Hawaiian people
- 1787 – The first male arrived in the Pacific Northwest from Hawaii
- 1811 – Hawaiian seamen begin intermarrying with Native American females along the Pacific shore, eventually expanding their presence from Oregon to Alaska
- 1839 – John Suter recruits Hawaiian men to travel with him to California
- 1845 – Hawaiians employed by Fort Vancouver, with some marrying Native American women
Conclusions
It’s without question that James Revels was Native American very early in the settlement of the Delmarva Peninsula, now Accomack County, Virginia, but his common ancestor with Filipino men 5100 years ago precludes his direct paternal ancestor’s presence in the Americas at that time. In other words, his Revel male ancestor did not arrive in the Beringian indigenous migration 12,000-16,000 years ago. His ancestor likely arrived post-contact, based on a combination of both historical and genetic evidence.
Haplogroup O is not found in the Arctic Inuit nor the Na-Dene speakers, precluding a connection with either group, and has never been found in ancient DNA in the Americas.
Haplogroup O in the Revels lineage is most likely connected with the Spanish galleon trade with the Philippines and the early Spanish attempts to colonize the Americas.
The source of Haplogroup O in the Pacific Northwest group is likely found in the recruitment of Hawaiian men in the early/mid-1800s.
The Mexican Haplogroup O group likely originated with the Manilla/Mexico Spanish galleon trade.
The source of the Blount Haplogroup O remains uncertain, other than to say it originated in Thailand thousands of years ago and is also found in the UAE. The common Blount, UAE, and Thailand ancestor’s haplogroup dates to 336 CE, so they were all likely in or near Thailand at that date, about 1687 years ago.
What’s Next?
Science continuously evolves, revealing new details as we learn more, often clarifying or shifting our knowledge. Before the Discover tool provided haplogroup ages based on tests from men around the world, we didn’t have the necessary haplogroup origin and age data to understand the genesis of haplogroup O in the Americas. Now, we do, but there is invariably more to learn.
New evidence is always welcome and builds our knowledge base. Haplogroup O ancient DNA findings would be especially relevant and could further refine what we know, depending on the location, dates of the remains, who they match, and historical context.
Additional Big Y-700 tests of haplogroup O men, especially those with known genealogy or ancestor location, including Madagascar, would be very beneficial and allow the haplogroup formation dates to be further refined.
If you are a male with haplogroup O, please consider upgrading to the Big Y-700 test, here.
_____________________________________________________________
Follow DNAexplain on Facebook, here.
Share the Love!
You’re always welcome to forward articles or links to friends and share on social media.
If you haven’t already subscribed (it’s free,) you can receive an email whenever I publish by clicking the “follow” button on the main blog page, here.
You Can Help Keep This Blog Free
I receive a small contribution when you click on some of the links to vendors in my articles. This does NOT increase the price you pay but helps me to keep the lights on and this informational blog free for everyone. Please click on the links in the articles or to the vendors below if you are purchasing products or DNA testing.
Thank you so much.
DNA Purchases and Free Uploads
Genealogy Products and Services
My Book
Genealogy Books
Genealogy Research
Like this:
Like Loading...