This is the fifth article in our series of articles about searching for unknown close family members, specifically; parents, grandparents, or siblings. However, these same techniques can be applied by genealogists to identify ancestors further back in time as well.
- I introduced the “In Search of” series in the article, DNA: In Search of…New Series Launches.
- In the second article, DNA: In Search of…What Do You Mean I’m Not Related to My Family? – and What Comes Next? we discussed the discovery that something was amiss when you don’t match a family member that you expect to match, then how to make sure a vial or upload mix-up didn’t happen. Next, I covered the basics of the four kinds of DNA tests you’ll be able to use to solve your mystery.
- In the third article, In Search of…Vendor Features, Strengths, and Testing Strategies, we discussed testing goals and strategies, including testing with and uploading to multiple autosomal DNA vendors, Y DNA, and mitochondrial DNA testing. We reviewed the vendor’s strengths and the benefits of combining vendor information and resources.
- In the fourth article, DNA: In Search of…Signs of Endogamy, we discussed the signs of endogamy and various ways to determine if you or your recent ancestors descend from an endogamous population.
Please note that if a family member has tested and you do NOT see their results, ask them to verify that they have chosen to allow matching and for other people to view them in their match list. That process varies at different vendors.
You can also ask if they can see you in their results.
All Parties Need to Test
Searching for unknown siblings isn’t exactly searching, because to find them, they, themselves, or their descendant(s) must have taken a DNA test at the same vendor where you tested or uploaded a DNA file.
You may know through any variety of methods that they exist, or might exist, but if they don’t take a DNA test, you can’t find them using DNA. This might sound obvious, but I see people commenting and not realizing that the other sibling(s) must test too – and they may not have.
My first questions when someone comments in this vein are:
- Whether or not they are positive their sibling actually tested, meaning actually sent the test in to the vendor, and it was received by the testing company. You’d be surprised how many tests are living in permanent residence on someone’s countertop until it gets pushed into the drawer and forgotten about.
- If the person has confirmed that their sibling has results posted. They may have returned their test, but the results aren’t ready yet or there was a problem.
- AND that both people have authorized matching and sharing of results. Don’t hesitate to reach out to your vendor’s customer care if you need help with this.
Sibling Scenarios
The most common sibling scenarios are when one of two things happens:
- A known sibling tests, only to discover that they don’t match you in the full sibling range, or not at all, when you expected they would
- You discover a surprise match in the full or half-sibling range
Let’s talk about these scenarios and how to determine:
- If someone is a sibling
- If they are a full or half-sibling
- If a half-sibling, if they descend from your mother or father
As with everything else genetic, we’ll be gathering and analyzing different pieces of evidence along the way.
Full and Half-Siblings
Just to make sure we are all on the same page:
- A full sibling is someone who shares both parents with you.
- A half-sibling is someone who shares one parent with you, but not the other parent.
- A step-sibling is someone who shares no biological parents with you. This situation occurs when your parent marries their parent, after you are both born, and their parent becomes your step-parent. You share neither of your biological parents with a step-sibling, so you share no DNA and will not show up on each other’s match lists.
- A three-quarters sibling is someone with whom you share one parent, but two siblings are the other parent. For example, you share the same mother, but one brother fathered you, and your father’s brother fathered your sibling. Yes, this can get very messy and is almost impossible for a non-professional to sort through, if even then. (This is not a solicitation. I do not take private clients.) We will not be addressing this situation specifically.
Caution

With any search for unknown relatives, you have no way of knowing what you will find.
In one’s mind, there are happy reunions, but you may experience something entirely different. Humans are human. Their stories are not always happy or rosy. They may have made mistakes they regret. Or they may have no regrets about anything.
Your sibling may not know about you or the situation under which you, or they, were born. Some women were victims of assault and violence, which is both humiliating and embarrassing. I wrote about difficult situations, here.
Your sibling or close family member may not be receptive to either you, your message, or even your existence. Just be prepared, because the seeking journey may not be pain-free for you or others, and may not culminate with or include happy reunions.
On the other hand, it may.
Please step back and ponder a bit about the journey you are about to undertake and the possible people that may be affected, and how. This box, once opened, cannot be closed again. Be sure you are prepared.
On the other hand, sometimes that box lid pops off, and the information simply falls in your lap one day when you open your match list, and you find yourself sitting there, in shock, staring at a match, trying to figure out what it all means.
Congratulations, You Have a Sibling!
This might not be exactly what runs through your mind when you see that you have a very close match that you weren’t expecting.

The first two things I recommend when making this sort of discovery, after a few deep breaths, a walk, and a cup of tea, are:
- Viewing what the vendor says
- Using the DNAPainter Shared cM Relationship Chart
Let’s start with DNAPainter.
DNAPainter
DNAPainter provides a relationship chart, here, based on the values from the Shared cM Project.

You can either enter a cM amount or a percentage of shared DNA. I prefer the cM amount, but it doesn’t really matter.
I’ll enter 2241 cM from a known half-sibling match. To enter a percent, click on the green “enter %.”

As you can see, statistically speaking, this person is slightly more likely to be a half-sibling than they are to be a full sibling. In reality, they could be either.
Looking at the chart below, DNAPainter highlights the possible relationships from the perspective of “Self.”

The average of all the self-reported relationships is shown, on top, so 2613 for a full sibling. The range is shown below, so 1613-3488 for a full sibling.
In this case, there are several possibilities for two people who share 2241 cM of DNA.
I happen to know that these two people are half-siblings, but if I didn’t, it would be impossible to tell from this information alone.
The cM range for full siblings is 1613-3488, and the cM range for half-siblings is 1160-2436.
- The lower part of the matching range, from 1160-1613 cM is only found in half-siblings.
- The portion of the range from 1613-2436 cM can be either half or full siblings.
- The upper part of the range, from 2436-3488 cM is only found in full siblings.
If your results fall into the center portion of the range, you’re going to need to utilize other tools. Fortunately, we have several.
If you’ve discovered something unexpected, you’ll want to verify using these tools, regardless. Use every tool available. Ranges are not foolproof, and the upper and lower 10% of the responses were removed as outliers. You can read more about the shared cM Project, here and here.
Furthermore, people may be reporting some half-sibling relationships as full sibling relationships, because they don’t expect to be half-siblings, so the ranges may be somewhat “off.”
Relationship Probability Calculator
Third-party matching database, GEDmatch, provides a Relationship Probability Calculator tool that is based on statistical probability methods without compiled user input. Both tools are free, and while I haven’t compared every value, both seem to be reasonably accurate, although they do vary somewhat, especially at the outer ends of the ranges.

When dealing with sibling matches, if you are in all four databases, GEDmatch is a secondary resource, but I will include GEDmatch when they have a unique tool as well as in the summary table. Some of your matches may be willing to upload to GEDmatch if the vendor where you match doesn’t provide everything you need and GEDmatch has a supplemental offering.
Next, let’s look at what the vendors say about sibling matches.
Vendors
Each of the major vendors reports sibling relationships in a slightly different way.
Sibling Matches at Ancestry
Ancestry reports sibling relationships as Sister or Brother, but they don’t say half or full.

If you click on the cM portion of the link, you’ll see additional detail, below

Ancestry tells you that the possible relationships are 100% “Sibling.” The only way to discern the difference between full and half is by what’s next.

If the ONLY relationship shown is Sibling at 100%, that can be interpreted to mean this person is a full sibling, and that a half-sibling or other relationship is NOT a possibility.
Ancestry never stipulates full or half.
The following relationship is a half-sibling at Ancestry.

Ancestry identifies that possible range of relationships as “Close Family to First Cousin” because of the overlaps we saw in the DNAPainter chart.

Clicking through shows that there is a range of possible relationships, and Ancestry is 100% sure the relationship is one of those.

DNAPainter agrees with Ancestry except includes the full-sibling relationship as a possibility for 1826 cM.
Sibling Matches at 23andMe
23andMe does identify full versus half-siblings.

DNAPainter disagrees with 23andMe and claims that anyone who shares 46.2% of their DNA is a parent/child.

However, look at the fine print. 23andMe counts differently than any of the other vendors, and DNAPainter relies on the Shared cM Project, which relies on testers entering known relationship matching information. Therefore, at any other vendor, DNAPainter is probably exactly right.
Before we understand how 23andMe counts, we need to understand about half versus fully identical segments.
To determine half or full siblings, 23andMe compares two things:
- The amount of shared matching DNA between two people
- Fully Identical Regions (FIR) of DNA compared to Half Identical Regions (HIR) of DNA to determine if any of your DNA is fully identical, meaning some pieces of you and your sibling’s DNA is exactly the same on both your maternal and paternal chromosomes.
Here’s an example on any chromosome – I’ve randomly selected chromosome 12. Which chromosome doesn’t matter, except for the X, which is different.

Your match isn’t broken out by maternal and paternal sides. You would simply see, on the chromosome browser, that you and your sibling match at these locations, above.
In reality, though, you have two copies of each chromosome, one from Mom and one from Dad, and so does your sibling.
In this example, Mom’s chromosome is visualized on top, and Dad’s is on the bottom, below, but as a tester, you don’t know that. All you know is that you match your sibling on all of those blue areas, above.

However, what’s actually happening in this example is that you are matching your sibling on parts of your mother’s chromosome and parts of your father’s chromosome, shown above as green areas
23andMe looks at both copies of your chromosome, the one you inherited from Mom, on top, and Dad, on the bottom, to see if you match your sibling on BOTH your mother’s and your father’s chromosomes in that location.
I’ve boxed the green matching areas in purple where you match your sibling fully, on both parents’ chromosomes.
If you and your sibling share both parents, you will share significant amounts of the same DNA on both copies of the same chromosomes, meaning maternal and paternal. In other words, full siblings share some purple fully identical regions (FIR) of DNA with each other, while half-siblings do not (unless they are also otherwise related) because half-siblings only share one parent with each other. Their DNA can’t be fully identical because they have a different parent that contributed the other copy of their chromosome.
|
Total Shared DNA |
Fully Identical DNA from Both Parents |
| Full Siblings |
~50% |
~25% |
| Half Siblings |
~25% |
0 |
- Full siblings are expected to share about 50% of the same DNA. In other words, their DNA will match at that location. That’s all the green boxed locations, above.
- Full siblings are expected to share about 25% of the same DNA from BOTH parents at the same location on BOTH copies of their chromosomes. These are fully identical regions and are boxed in purple, above.
You’ll find fully identical segments about 25% of the time in full siblings, but you won’t find fully identical segments in half-siblings. Please note that there are exceptions for ¾ siblings and endogamous populations.

You can view each match at 23andMe to see if you have any completely identical regions, shown in dark purple in the top comparison of full siblings. Half siblings are shown in the second example, with less total matching DNA and no FIR or completely identical regions.
Please note that your matching amount of DNA will probably be higher at 23andMe than at other companies because:
- 23andMe includes the X chromosome in the match totals
- 23andMe counts fully identical matching regions twice. For full siblings, that’s an additional 25%
Therefore, a full sibling with an X match will have a higher total cM at 23andMe than the same siblings elsewhere because not only is the X added into the total, the FIR match region is added a second time too.
Fully Identical Regions (FIR) and Half Identical Regions (HIR) at GEDmatch
At GEDMatch, you can compare two people to each other, with an option to display the matching information and a painted graphic for each chromosome that includes FIR and HIR.
If you need to know if you and a match share fully identical regions and you haven’t tested at 23andMe, you can both upload your DNA data file to GEDmatch and use their One to One Autosomal DNA Comparison.

On the following page, simply enter both kit numbers and accept the defaults, making sure you have selected one of the graphics options.

While GEDmatch doesn’t specifically tell you whether someone is a full or half sibling, you can garner additional information about the relationship based on the graphic at GEDmatch.
GEDMatch shows both half and fully identical regions.
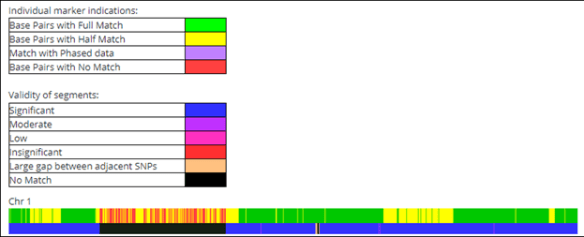
The above match is between two full siblings using a 7 cM threshold. The blue on the bottom bar indicates a match of 7 cM or larger. Black means no match.
The green regions in the top bar indicate places where these two people carry the same DNA on both copies of their chromosome 1. This means that both people inherited the same DNA from BOTH parents on the green segments.
In the yellow regions, the siblings inherited the same DNA from ONE parent, but different DNA in that region from the other parent. They do match each other, just on one of their chromosomes, not both.
Without a tool like this to differentiate between HIR and FIR, you can’t tell if you’re matching someone on one copy of your chromosome, or on both copies.
In the areas marked with red on top, which corresponds to the black on the bottom band, these two siblings don’t match each other because they inherited different DNA from both parents in that region. The yellow in that region is too scattered to be significant.
Full siblings generally share a significant amount of FIR, or fully identical regions of DNA – about 25%.
Half siblings will share NO significant amount of FIR, although some will be FIR on very small, scattered green segments simply by chance, as you can see in the example, below.

This half-sibling match shares no segments large enough to be a match (7 cM) in the black section. In the blue matching section, only a few small green fragments of DNA match fully, which, based on the rest of that matching segment, must be identical by chance or misreads. There are no significant contiguous segments of fully identical DNA.
When dealing with full or half-siblings, you’re not interested in small, scattered segments of fully identical regions, like those green snippets on chromosome 6, but in large contiguous sections of matching DNA like the chromosome 1 example.
GEDmatch can help when you match when a vendor does not provide FIR/HIR information, and you need additional assistance.
Next, let’s look at full and half-siblings at FamilyTreeDNA
Sibling Matches at FamilyTreeDNA
FamilyTreeDNA does identify full siblings.

Relationships other than full siblings are indicated by a range. The two individuals below are both half-sibling matches to the tester.
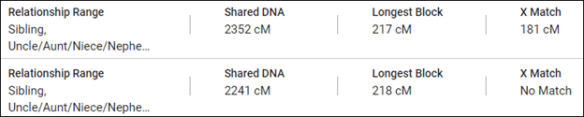
The full range when mousing over the relationship ranges is shown below.

DNAPainter agrees except also gives full siblings as an option for the two half-siblings.
FamilyTreeDNA also tells you if you have an X match and the size of your X match.
We will talk about X matching in a minute, which, when dealing with sibling identification, can turn out to be very important.
Sibling Matches at MyHeritage
MyHeritage indicates brother or sister for full siblings

MyHeritage provides other “Estimated relationships” for matches too small to be full siblings.

DNAPainter’s chart agrees with this classification, except adds additional relationship possibilities.

Be sure to review all of the information provided by each vendor for close relationships.
View Close Known Relationships
The next easiest step to take is to compare your full or half-sibling match to known close family members from your maternal and paternal sides, respectively. The closer the family members, the better.
It’s often not possible to determine if someone is a half sibling or a full sibling by centiMorgans (cMs) alone, especially if you’re searching for unknown family members.
Let’s start with the simplest situation first.
Let’s say both of your parents have tested, and of course, you match both of them as parents.
Your new “very close match” is in the sibling range.
The first thing to do at each vendor is to utilize that vendor’s shared matches tool and see whether your new match matches one parent, or both.
Here’s an example.
Close Relationships at FamilyTreeDNA
This person has a full sibling match, but let’s say they don’t know who this is and wants to see if their new sibling matches one or both of their parents.

Select the match by checking the box to the left of the match name, then click on the little two-person icon at far right, which shows “In Common” matches

You can see on the resulting shared match list that both of the tester’s parents are shown on the shared match list.
Now let’s make this a little more difficult.
No Parents, No Problem
Let’s say neither of your parents has tested.
If you know who your family is and can identify your matches, you can see if the sibling you match matches other close relatives on both or either side of your family.
You’ll want to view shared matches with your closest known match on both sides of your tree, beginning with the closest first. Aunts, uncles, first cousins, etc.
You will match all of your family members through second cousins, and 90% of your third cousins. You can view additional relationship percentages in the article, How Much of Them is in You?.
I recommend, for this matching purpose, to utilize 2nd cousins and closer. That way you know for sure if you don’t share them as a match with your sibling, it’s because the sibling is not related on that side of the family, not because they simply don’t share any DNA due to their distance.

In this example, you have three sibling matches. Based on your and their matches to the same known first and second cousins, you can see that:
- Sibling 1 is your full sibling, because you both match the same maternal and paternal first and second cousins
- Sibling 2 is your paternal half-sibling because you both match paternal second cousins and closer, but not maternal cousins.
- Sibling 3 is your maternal half-sibling because you both match maternal second cousins and closer, but not paternal cousins.
Close Relationships at Ancestry
Neither of my parents have tested, but my first cousin on my mother’s side has. Let’s say I have a suspected sibling or half-sibling match, so I click on the match’s name, then on Shared Matches.

Sure enough, my new match also matches my first cousin that I’ve labeled as “on my mother’s side.”
If my new match in the sibling range also matches my second cousins or closer on my father’s side, the new match is a full sibling, not a half-sibling.
Close Relationships at MyHeritage
Comparing my closest match provided a real surprise. I wonder if I’ve found a half-sibling to my mother.

Now, THIS is interesting.
Hmmm. More research is needed, beginning with the age of my match. MyHeritage provides ages if the MyHeritage member authorizes that information to be shared.
Close Relationships at 23andMe
Under DNA Relatives, click on your suspected sibling match, then scroll down and select “Find Relatives in Common.”

The Relatives in Common list shows people that match both of you.
The first common match is very close and a similar relationship to my closest match on my father’s side. This would be expected of a sibling. I have no common matches with this match to anyone on my mother’s side, so they are only related on my father’s side. Therefore they are a paternal half-sibling, not a full sibling.

More Tools Are Available
Hopefully, by now, you’ve been able to determine if your mystery match is a sibling, and if so, if they are a half or full sibling, and through which parent.
We have some additional tools that are relevant and can be very informative in some circumstances. I suggest utilizing these tools, even if you think you know the answer.
In this type of situation, there’s no such thing as too much information.
X Matching
X matching, or lack thereof, may help you determine how you are related to someone.
There are two types of autosomal DNA. The X chromosome versus chromosomes 1-22. The X chromosome (number 23) has a unique inheritance path that distinguishes it from your other chromosomes.
The X chromosome inheritance path also differs between men and women.

Here’s my pedigree chart in fan form, highlighting the ancestors who may have contributed a portion of their X chromosome to me. In the closest generation, this shows that I inherited an X chromosome from both of my parents, and who in each of their lines could have contributed an X to them.
The white or uncolored positions, meaning ancestors, cannot contribute any portion of an X chromosome to me based on how the X chromosome is inherited.
You’ll notice that my father inherited none of his X chromosome from any of his paternal ancestors, so of course, I can’t inherit what he didn’t inherit. There are a very limited number of ancestors on my father’s side whom I can inherit any portion of an X chromosome from.

Men receive their Y chromosome from their fathers, so men ONLY receive an X chromosome from their mother.
Therefore, men MUST pass their mother’s X chromosome on to their female offspring because they don’t have any other copy of the X chromosome to pass on.
Men pass no X chromosome to sons.
We don’t need to worry about a full fan chart when dealing with siblings and half-siblings.
We only need to be concerned with the testers plus one generation (parents) when utilizing the X chromosome in sibling situations.

These two female Disney Princesses, above, are full siblings, and both inherited an X chromosome from BOTH their mother and father. However, their father only has one X (red) chromosome to give them, so the two females MUST match on the entire red X chromosome from their father.
Their mother has two X chromosomes, green and black, to contribute – one from each of her parents.
The full siblings, Melody, and Cinderella:
- May have inherited some portion of the same green and black X chromosomes from their mother, so they are partial matches on their mother’s X chromosome.
- May have inherited the exact same full X chromosome from their mother (both inherited the entire green or both inherited the entire black), so they match fully on their mother’s X chromosome.
- May have inherited the opposite X from different maternal grandparents. One inherited the entire green X and one inherited the entire black X, so they don’t match on their mother’s X chromosome.
Now, let’s look at Cinderella, who matches Henry.

This female and male full sibling match can’t share an X chromosome on the father’s side, because the male’s father doesn’t contribute an X chromosome to him. The son, Henry, inherited a Y chromosome instead from his father, which is what made them males.
Therefore, if a male and female match on the X chromosome, it MUST be through HIS mother, but could be through either of her parents. In a sibling situation, an X match between a male and female always indicates the mother.
In the example above, the two people share both of their mother’s X chromosomes, so are definitely (at least) maternally related. They could be full siblings, but we can’t determine that by the X chromosome in this situation, with males.
However, if the male matches the female on HER father’s X chromosome, there a different message, example below.

You can see that the male is related to the female on her father’s side, where she inherited the entire magenta X chromosome. The male inherited a portion of the magenta X chromosome from his mother, so these two people do have an X match. However, he matches on his mother’s side, and she matches on her father’s side, so that’s clearly not the same parent.
- These people CAN NOT be full siblings because they don’t match on HER mother’s side too, which would also be his mother’s side if they were full siblings.
- They cannot be maternal half-siblings because their X DNA only matches on her father’s side, but they wouldn’t know that unless she knew which side was which based on share matches.
- They cannot be paternal half-siblings because he does not have an X chromosome from his father.
They could, however, be uncle/aunt-niece/nephew or first cousins on his mother’s side and her father’s side. (Yes, you’re definitely going to have to read this again if you ever need male-female X matching.)
Now, let’s look at X chromosome matching between two males. It’s a lot less complicated and much more succinct.

Neither male has inherited an X chromosome from their father, so if two males DO match on the X, it MUST be through their mother. In terms of siblings, this would mean they share the same mother.
However, there is one slight twist. In the above example, you can see that the men inherited a different proportion of the green and black X chromosomes from their common mother. However, it is possible that the mother could contribute her entire green X chromosome to one son, Justin in this example, and her entire black X chromosome to Henry.

Therefore, even though Henry and Justin DO share a mother, their X chromosome would NOT match in this scenario. This is rare but does occasionally happen.
Based on the above examples, the X chromosome may be relevant in the identification of full or half siblings based on the sexes of the two people who otherwise match at a level indicating a full or half-sibling relationship.
Here’s a summary chart for sibling X matching.
| X Match |
Female |
Male |
| Female |
Will match on shared father’s full X chromosome, mother’s X is the same rules as chromosomes 1-22 |
Match through male’s mother, but either of female’s parents. If the X match is not through the female’s mother, they are not full siblings nor maternal half-siblings. They cannot have an X match through the male’s father. They are either full or half-siblings through their mother if they match on both of their mother’s side. If they match on his mother’s side, and her father’s side, they are not siblings but could be otherwise closely related. |
| Male |
Match through male’s mother, but either of female’s parents. If the X match is not through the female’s mother, they are not full siblings nor maternal half-siblings. They cannot have an X match through the male’s father. They are either full or half-siblings through their mother if they match on both or their mother’s side. If they match on his mother’s side, and her father’s side, they are not siblings but could be otherwise closely related. |
Both males are related on their mother’s side – either full or half-siblings. |
Here’s the information presented in a different way.
DOES match X summary:
- If a male DOES match a female on the X, he IS related to her through HIS mother’s side, but could match her on her mother or father’s side. If their match is not through her mother, then they are not full siblings nor maternal half-siblings. They cannot match through his father, so they cannot be paternal half-siblings.
- If a female DOES match a female on the X, they could be related on either side and could be full or half-siblings.
- If a male DOES match a male on the X, they ARE both related through their mother. They may also be related on their father’s side, but the X does not inform us of that.
Does NOT match X summary:
- If a male does NOT match a female on the X, they are NOT related through HIS mother and are neither full siblings nor maternal half-siblings. Since a male does not have an X chromosome from his father, they cannot be paternal half-siblings based on an X match.
- If a male does NOT match a male, they do NOT share a mother.
- If a female does NOT match another female on the X, they are NOT full siblings and are NOT half-siblings on their paternal side. Their father only has one X chromosome, and he would have given the same X to both daughters.
Of the four autosomal vendors, only 23andMe and FamilyTreeDNA report X chromosome results and matching, although the other two vendors, MyHeritage and Ancestry, include the X in their DNA download file so you can find X matches with those files at either FamilyTreeDNA or GEDMatch if your match has or will upload their file to either of those vendors. I wrote step-by-step detailed download/upload instructions, here.
X Matching at FamilyTreeDNA

In this example from FamilyTreeDNA, the female tester has discovered two half-sibling matches, both through her father. In the first scenario, she matches a female on the full X chromosome (181 cM). She and her half-sibling MUST share their father’s entire X chromosome because he only had one X, from his mother, to contribute to both of his daughters.
In the second match to a male half-sibling, our female tester shares NO X match because her father did not contribute an X chromosome to his son.
If we didn’t know which parents these half-sibling matches were through, we can infer from the X matching alone that the male is probably NOT through the mother.
Then by comparing shared matches with each sibling, Advanced Matches, or viewing the match Matrix, we can determine if the siblings match each other and are from the same or different sides of the family.

Under Additional Tests and Tools, Advanced Matching, FamilyTreeDNA provides an additional tool that can show only X matches combined with relationships.

Of course, you’ll need to view shared matches to see which people match the mother and/or match the father.
To see who matches each other, you’ll need to use the Matrix tool.

At FamilyTreeDNA, the Matrix, located under Autosomal DNA Results and Tools, allows you to select your matches to see if they also match each other. If you have known half-siblings, or close relatives, this is another way to view relationships.

Here’s an example using my father and two paternal half-siblings. We can see that the half-siblings also match each other, so they are (at least) half-siblings on the paternal side too.
If they also matched my mother, we would be full siblings, of course.
Next, let’s use Y DNA and mitochondrial DNA.
Y DNA and Mitochondrial DNA

In addition to autosomal DNA, we can utilize Y DNA and mitochondrial DNA (mtDNA) in some cases to identify siblings or to narrow or eliminate relationship possibilities.
Given that Y DNA and mitochondrial DNA both have distinctive inheritance paths, full and half-siblings will, or will not, match under various circumstances.
Y DNA
Y DNA is passed intact from father to son, meaning it’s not admixed with any of the mother’s DNA. Daughters do not inherit Y DNA from their father, so Y DNA is only useful for male-to-male comparisons.
Two types of Y DNA are used for genealogy, STR markers for matching, and haplogroups, and both are equally powerful in slightly different ways.
Y DNA at FamilyTreeDNA
Men can order either 37 or 111 STR marker tests, or the BIg Y which provides more than 700 markers and more. FamilyTreeDNA is the only one of the vendors to offer Y DNA testing that includes STR markers and matching between men.
Men who order these tests will be compared for matching on either 37, 111 or 700 STR markers in addition to SNP markers used for haplogroup identification and assignment.
Fathers will certainly match their sons, and paternal line brothers will match each other, but they will also match people more distantly related.
However, if two men are NOT either full or half siblings on the paternal side, they won’t match at 111 markers.
If two men DON’T match, especially at high marker levels, they likely aren’t siblings. The word “likely” is in there because, very occasionally, a large deletion occurs that prevents STR matching, especially at lower levels.
Additionally, men who take the 37 or 111 marker test also receive an estimated haplogroup at a high level for free, without any additional testing.
However, if men take the Big Y-700 test, they not only will (or won’t) match on up to 700 STR markers, they will also receive a VERY refined haplogroup via SNP marker testing that is often even more sensitive in terms of matching than STR markers. Between these two types of markers, Y DNA testing can place men very granularly in relation to other men.
Men can match in two ways on Y DNA, and the results are very enlightening.

If two men match on BOTH their most refined haplogroup (Big Y test) AND STR markers, they could certainly be siblings or father/son. They could also be related on the same line for another reason, such as known or unknown cousins or closer relationships like uncle/nephew. Of course, Y DNA, in addition to autosomal matching, is a powerful combination.
Conversely, if two men don’t have a similar or close haplogroup, they are not a father and son or paternal line siblings.
FamilyTreeDNA offers both inexpensive entry-level testing (37 and 111 markers) and highly refined advanced testing of most of the Y chromosome (Big Y-700), so haplogroup assignments can vary widely based on the test you take. This makes haplogroup matching and interpretation a bit more complex.
For example, haplogroups R-M269 and I-BY14000 are not related in thousands of years. One is haplogroup R, and one is haplogroup I – completely different branches of the Y DNA tree. These two men won’t match on STR markers or their haplogroup.
However, because FamilyTreeDNA provides over 50,000 different haplogroups, or tree branches, for Big Y testers, and they provide VERY granular matching, two father/son or sibling males who have BOTH tested at the Big Y-700 level will have either the exact same haplogroup, or at most, one branch difference on the tree if a mutation occurred between father and son.
If both men have NOT tested at the Big Y-700 level, their haplogroups will be on the same branch. For example, a man who has only taken a 37/111 marker STR test may be estimated at R-M269, which is certainly accurate as far as it goes.
His sibling who has taken a Big Y test will be many branches further downstream on the tree – but on the same large haplogroup R-M269 branch. It’s essential to pay attention to which tests a Y DNA match has taken when analyzing the match.
The beauty of the two kinds of tests is that even if one haplogroup is very general due to no Big Y test, their STR markers should still match. It’s just that sometimes this means that one hand is tied behind your back.
Y DNA matching alone can eliminate the possibility of a direct paternal line connection, but it cannot prove siblingship or paternity alone – not without additional information.

The Advanced Matching tool will provide a list of matches in all categories selected – in this case, both the 111 markers and the Family Finder test. You can see that one of these men is the father of the tester, and one is the full sibling.
You can view haplogroup assignments on the public Y DNA tree, here. I wrote about using the public tree, here.
In addition, recently, FamilyTreeDNA launched the new Y DNA Discover tool, which explains more about haplogroups, including their ages and other fun facts like migration paths along with notable and ancient connections. I wrote about using the Discover tool, here.
Y DNA at 23andMe
Testers receive a base haplogroup with their autosomal test. 23andMe tests a limited number of Y DNA SNP locations, but they don’t test many, and they don’t test STR markers, so there is no Y DNA matching and no refined haplogroups.
You can view the haplogroups of your matches. If your male sibling match does NOT share the same haplogroup, the two men are not paternal line siblings. If two men DO share the same haplogroup, they MIGHT be paternal siblings. They also might not.
Again, autosomal close matching plus haplogroup comparisons include or exclude paternal side siblings for males.

Paternal side siblings at 23andMe share the same haplogroup, but so do many other people. These two men could be siblings. The haplogroups don’t exclude that possibility. If the haplogroups were different, that would exclude being either full or paternal half-siblings.
Men can also compare their mitochondrial DNA to eliminate a maternal relationship.

These men are not full siblings or maternal half-siblings. We know, unquestionably, because their mitochondrial haplogroups don’t match.
23andMe also constructs a genetic tree, but often struggles with close relative placement, especially when half-relationships are involved. I do not recommend relying on the genetic tree in this circumstance.
Mitochondrial DNA
Mitochondrial DNA is passed from mothers to all of their children, but only females pass it on. If two people, males or females, don’t match on their mitochondrial DNA test, with a couple of possible exceptions, they are NOT full siblings, and they are NOT maternal half-siblings.
Mitochondrial DNA at 23andMe
23andMe provides limited, base mitochondrial haplogroups, but no matching. If two people don’t have the same haplogroup at 23andMe, they aren’t full or maternal siblings, as illustrated above.
Mitochondrial DNA at FamilyTreeDNA
FamilyTreeDNA provides both mitochondrial matching AND a much more refined haplogroup. The full sequence test (mtFull), the only version sold today, is essential for reliable comparisons.
Full siblings or maternal half-siblings will always share the same haplogroup, regardless of their sex.
Generally, a full sibling or maternal half-sibling match will match exactly at the full mitochondrial sequence (FMS) level with a genetic distance of zero, meaning fully matching and no mismatching mutations.

There are rare instances where maternal siblings or even mothers and children do not match exactly, meaning they have a genetic distance of greater than 0, because of a mutation called a heteroplasmy.
I wrote about heteroplasmies, here.
Like Y DNA, mitochondrial DNA cannot identify a sibling or parental relationship without additional evidence, but it can exclude one, and it can also provide much-needed evidence in conjunction with autosomal matching. The great news is that unlike Y DNA, everyone has mitochondrial DNA and it comes directly from their mother.
Once again, FamilyTreeDNA’s Advanced Matching tool provides a list of people who match you on both your mitochondrial DNA test and the Family Finder autosomal test, including transfers/uploads, and provides a relationship.

You can see that our tester matches both a full sibling and their mother. Of course, a parent/child match could mean that our tester is a female and one of her children, of either sex, has tested.
Below is an example of a parent-child match that has experienced a heteroplasmy.

Based on the comparison of both the mitochondrial DNA test, plus the autosomal Family Finder test, you can verify that this is a close family relationship.
You can also eliminate potential relationships based on the mitochondrial DNA inheritance path. The mitochondrial DNA of full siblings and maternal half-siblings will always match at the full sequence and haplogroup level, and paternal half-siblings will never match. If paternal half-siblings do match, it’s happenstance or because of a different reason.
Sibling Summary and Checklist
I’ve created a quick reference checklist for you to use when attempting to determine whether or not a match is a sibling, and, if so, whether they are half or full siblings. Of course, these tools are in addition to the DNAPainter Shared cM Tool and GEDmatch’s Relationship Predictor Calculator.
|
FamilyTreeDNA |
Ancestry |
23andMe |
MyHeritage |
GEDmatch |
| Matching |
Yes |
Yes |
Yes |
Yes |
Yes |
| Shared Matches |
Yes – In Common With |
Yes – Shared Matches |
Yes – Relatives in Common |
Yes – Review DNA Match |
Yes – People who match both or 1 of 2 kits |
| Relationship Between Shared Matches |
No |
No |
No |
Yes, under shared match |
No |
| Matches Match Each Other* |
Yes, Matrix |
No |
Yes, under “View DNA details,” then, “compare with more relatives” |
Partly, through triangulation |
Yes, can match any kits |
| Full Siblings |
Yes |
Sibling, implies full |
Yes |
Brother, Sister, means full |
No |
| Half Siblings |
Sibling, Uncle/Aunt-Niece/Nephew, Grandparent-Grandchild |
Close Family – 1C |
Yes |
Half sibling, aunt/uncle-niece-nephew |
No |
| Fully Identical Regions (FIR) |
No |
No |
Yes |
No |
Yes |
| Half Identical Regions (HIR) |
No |
No |
Yes |
No |
Yes |
| X matching |
Yes |
No |
Yes |
No |
Yes |
| Unusual Reporting or Anomalies |
No |
No, Timber is not used on close relationships |
X match added into total, FIR added twice |
No |
Matching amount can vary from vendors |
| Y DNA |
Yes, STRs, refined haplogroups, matching |
No |
High-level haplogroup only, no matching |
No |
No, only if tester enters haplogroup manually |
| Mitochondrial DNA |
Yes, full sequence, matching, refined haplogroup |
No |
High-level haplogroup only, no matching |
No |
No, only if tester enters haplogroup manually |
| Combined Tools (Autosomal, X, Y, mtDNA) |
Yes |
No |
No |
No |
No |
*Autoclusters through Genetic Affairs show cluster relationships of matches to the tester and to each other, but not all matches are included, including close matches. While this is a great tool, it’s not relevant for determining close and sibling relationships. See the article, AutoClustering by Genetic Affairs, here.
Additional Resources
Some of you may be wondering how endogamy affects sibling numbers.
Endogamy makes almost everything a little more complex. I wrote about endogamy and various ways to determine if you have an endogamous heritage, here.
Please note that half-siblings with high cM matches also fall into the range of full siblings (1613-3488), with or without endogamy. This may be, but is not always, especially pronounced in endogamous groups.
As another resource, I wrote an earlier article, Full or Half Siblings, here, that includes some different examples.
Strategy
You have a lot of quills in your quiver now, and I wish you the best if you’re trying to unravel a siblingship mystery.
You may not know who your biological family is, or maybe your sibling doesn’t know who their family is, but perhaps your close relatives know who their family is and can help. Remember, the situation that has revealed itself may be a shock to everyone involved.
Above all, be kind and take things slow. If your unexpected sibling match becomes frightened or overwhelmed, they may simply check out and either delete their DNA results altogether or block you. They may have that reaction before you have a chance to do anything.
Because of that possibility, I recommend performing your analysis quickly, along with taking relevant screenshots before reaching out so you will at least have that much information to work with, just in case things go belly up.
When you’re ready to make contact, I suggest beginning by sending a friendly, short, message saying that you’ve noticed that you have a close match (don’t say sibling) and asking what they know about their family genealogy – maybe ask who their grandparents are or if they have family living in the area where you live. I recommend including a little bit of information about yourself, such as where you were born and are from.
I also refrain from using the word adoption (or similar) in the beginning or giving too much detailed information, because it sometimes frightens people, especially if they know or discover that there’s a painful or embarrassing family situation.
And, please, never, ever assume the worst of anyone or their motives. They may be sitting at their keyboard with the same shocked look on their face as you – especially if they have, or had, no idea. They may need space and time to reach a place of acceptance. There’s just nothing more emotionally boat-capsizing in your life than discovering intimate and personal details about your parents, one or both, especially if that discovery is disappointing and image-altering.
Or, conversely, your sibling may have been hoping and waiting just for you!
Take a deep breath and let me know how it goes!
Please feel free to share this article with anyone who could benefit.
_____________________________________________________________
Follow DNAexplain on Facebook, here or follow me on Twitter, here.
Share the Love!
You’re always welcome to forward articles or links to friends and share on social media.
If you haven’t already subscribed (it’s free,) you can receive an email whenever I publish by clicking the “follow” button on the main blog page, here.
You Can Help Keep This Blog Free
I receive a small contribution when you click on some of the links to vendors in my articles. This does NOT increase the price you pay but helps me to keep the lights on and this informational blog free for everyone. Please click on the links in the articles or to the vendors below if you are purchasing products or DNA testing.
Thank you so much.
DNA Purchases and Free Uploads
Genealogy Products and Services
My Book
Genealogy Books
Genealogy Research
Like this:
Like Loading...