There has been quite a bit of discussion in the last several weeks, both pro and con, about how to use small matching DNA segments in genetic genealogy. A couple of people are even of the opinion that small segments can’t be used at all, ever. Others are less certain and many of us are working our way through various scenarios. Evidence certainly exists that these segments can be utilized.
I’ve been writing foundation articles, in preparation for this article, for several weeks now. Recently, I wrote about how phasing works and determining IBD versus IBS matches and included guidelines for telling the difference between the different kinds of matches. If you haven’t read that article, it’s essential to understanding this article, so now would be a good time to read or review that article.
I followed that with a step by step article, Demystifying Autosomal DNA Matching, on how to do phasing and matching in combination with the guidelines about how to determine IBD (identical by descent) versus IBS (identical by chance) and identical by population matches when evaluating your own matches.
Now that we understand IBS, IBD, Phasing and how matching actually works on a case by case basis, let’s look at applying those same matching and IBS vs IBD guidelines to small data segments as well.
A Little History
So those of you who haven’t been following the discussion on various blogs and social media don’t feel like you’ve been dropped into the middle of a conversation with no context, let me catch you up.
On Thanksgiving Day, I published an article about identifying one of my ancestors, after many years of trying, Sarah Hickerson.
That article spurred debate, which is just fine when the debate is about the science, but it subsequently devolved into something less pleasant. There are some individuals with very strong opinions that utilizing small segments of DNA data can “never be done.”
I do not agree with that position. In fact, I strongly disagree and there are multiple cases with evidence to support small segments being both accurate and useful in specific types of genealogical situations. We’ll take a look at several.
I do agree that looking at small segment data out of context is useless. To the best of my knowledge, no genealogist begins with their smallest segments and tries to assemble them, working from the bottom up. We all begin with the largest segments, because they are the most useful and the closest connections in our tree, and work our way down. Generally, we only work with small segments when we have to – and there are times that’s all we have. So we need to establish guidelines and ways to know if those small segments are reliable or not. In other words, how can we draw conclusions and how much confidence can we put in those conclusions?
Ultimately, whether you choose to use or work with small segment data will be your own decision, based on your own circumstances. I simply wanted to understand what is possible and what is reasonable, both for my own genealogy and for my readers.
In my projects, I haven’t been using small segment data out of context, or randomly. In other words, I don’t just pick any two small segment matches and infer or decide that they are valid matches. Fortunately, by utilizing the IBD vs IBS guidelines, we have tools to differentiate IBD (Identical by Descent) segments from IBS (Identical by State) by chance segments and IBD/IBS by population for matching segments, both large and small.
Studying small segment data is the key to determining exactly how small segments can reasonably be utilized. This topic probably isn’t black or white, but shades of gray – and assuming the position that something can’t be done simply assures that it won’t be.
I would strongly encourage those involved and interested in this type of research to retain those small segments, work with them and begin to look for patterns. The only way we, as a community, are ever going to figure out how to work with small segments successfully and reliably is to, well, work with them.
Discussing the science and scenarios surrounding the usage of small data segments in various different situations is critical to seeing our way through the forest. If the answers were cast in concrete about how to do this, we wouldn’t be working through this publicly today.
Negative personal comments and inferences have no place in the scientific community. It discourages others from participating, and serves to stifle research and cooperation, not encourage it. I hope that civil scientific discussions and comparisons involving small segment data can move forward, with decorum, because they are critically needed in order to enhance our understanding, under varying circumstances, of how to utilize small segment data. As Judy Russell said, disagreeing doesn’t have to be disagreeable.
Two bloggers, Blaine Bettinger and CeCe Moore wrote articles following my Hickerson article. Blaine subsequently wrote a second article here. Felix Immanuel wrote articles here and here.
A few others have weighed in, in writing, as well although most commentary has been on Facebook. Israel Pickholtz, a professional genealogist and genetic consultant, stated on his blog, All My Foreparents, the following:
It is my nature to distrust rules that put everything into a single category and that’s how I feel about small segments. Sometimes they are meaningful and useful, sometimes not.
When I reconstructed my father’s DNA using Lazerus (described last week in Genes From My Father), I happily accepted all small segments of whatever size because those small segments were in the DNA of at least one of his children and at least one of his brother/sister/first cousin. If I have a particular small segment, I must have received it from my parents. If my father’s brother (or sister) has it as well, then it is eminently clear to me that I got it from my father and that it came to him and his brother from my grandfather. And it is not reasonable to say that a sliver of that small segment might have come from my mother, because my father’s people share it.
After seeing Israel’s commentary about Lazarus, I reconstructed the genome of both Roscoe and John Ferverda, brothers, which includes both large and small segments. Working with the Ferverda DNA further, I wrote an article, Just One Cousin, about matching between two siblings and a first cousin, which includes lots of small data segments, some of which were proven to triangulate, meaning they are genuine, and some which did not. There are lots more examples in the demystifying article, as well.
What Not To Do
Before we begin, I want to make it very clear that am not now, and never have, advocated that people utilize small data segments out of context of larger matching segments and/or at least suspected matching genealogy. For example, I have never implied or even hinted that anyone should go to GedMatch, do a “one to many” compare at 1 cM and then contact people informing them that they are related. Anyone who has extrapolated what I’ve written to mean that either simply did not understand or intentionally misinterpreted the articles.
Sarah Hickerson Revisited
If I thought Sarah Hickerson caused me a lot of heartburn in the decades before I found her, little did I know how much heartburn that discovery would cause.
Let’s go back to the Sarah Hickerson article that started the uproar over whether small data segments are useful at all.
In that article, I found I was a member of a new Ancestry DNA Circle for Charles Hickerson and Mary Lytle, the parents of Sarah Hickerson.

Because there are no tools at Ancestry to prove DNA connections, I hurried over to Family Tree DNA looking for any matches to Hickersons for myself and for my Vannoy cousins who also (potentially) descended from this couple. Much to my delight, I found several matches to Hickersons, in fact, more than 20 – a total of 614 rows of spreadsheet matches when I included all of my Vannoy cousins who potentially descend from this couple to their Hickerson matches. There were 64 matching clusters of segments, both small and large. Some matches were as large as 20cM with 6000 SNPs and more than 20 were over 10cM with from 1500 to 6000 SNPs. There were also hundreds of small segments that matched (and triangulated) as well.
By the time I added in a few more Vannoy cousins that we’ve since recruited, the spreadsheet is now up to 1093 rows and we have 52 Vannoy-Hickerson TRIANGULATED CLUSTERS utilizing only Family Tree DNA tools.
Triangulated DNA, found in 3 or more people at the same location who share a common ancestor is proven to be from that ancestor (or ancestral couple.) This is the commonly accepted gold standard of autosomal DNA triangulation within the industry.
Here’s just one example of a cluster of three people. Charlene and Buster are known (proven, triangulated) cousins and Barbara is a descendant of Charles Hickerson and Mary Lytle.

What more could you want?
Yes, I called this a match. As far as I’m concerned, it’s a confirmed ancestor. How much more confirmed can you get?
Some clusters have as many as 25 confirmed triangulated members.

Others took issue with this conclusion because it included small segment data. This seems like the perfect opportunity in which to take a look at how small segments do, or don’t stand up to scrutiny. So, let’s do just that. I also did the same type of matching comparison in a situation with 2 siblings and a known cousin, here.
To Trash…or Not To Trash
Some genetic genealogists discard small segments entirely, generally under either 5 or 7cM, which I find unfortunate for several reasons.
- If a person doesn’t work with small segments, they really can’t comment on the lack of results, and they’ll never have a success because the small segments will have been discarded.
- If a person doesn’t work with small segments, they will never notice any trends or matches that may have implications for their ancestry.
- If a person doesn’t work with small segments, they can’t contribute to the body of evidence for how to reasonably utilize these segments.
- If a person doesn’t work with small segments, they may well be throwing the baby out with the bathwater, but they’ll never know.
- They encourage others to do the same.
The Sarah Hickerson article was not meant as a proof article for anything – it was meant to be an article encouraging people to utilize genetic genealogy for not only finding their ancestor and proving known connections, but breaking down brick walls. It was pointing the way to how I found Sarah Hickerson. It was one of my 52 Ancestors Series, documenting my ancestors, not one of the specifically educational articles. This article is different.
If you are only interested in the low hanging fruit, meaning within the past 5 or 6 generations, and only proving your known pedigree, not finding new ancestors beyond that 5-6 generation level, then you can just stop reading now – and you can throw away your small segments. But if you want more, then keep reading, because we as a community need to work with small segment data in order to establish guidelines that work relative to utilizing small segments and identifying the small segments that can be useful, versus the ones that aren’t.
I do not believe for one minute that small segments are universally useless. As Israel said, if his family did not receive those segments from a common family member, then where did they all get those matching segments?
In fact, utilizing triangulated and proven DNA relationships within families is how adoptees piece together their family trees, piggybacking off of the work of people with known pedigrees that they match genetically. My assumption had been that the adoptee community utilized only large DNA segments, because the larger the matching segments, generally the closer in time the genealogy match – and theoretically the easier to find.
However, I discovered that I was wrong, and the adoptee community does in fact utilize small segments as well. Here’s one of the comments posted on my Chromosome Browser War blog article.
“Thanks for the well thought out article, Roberta, I have something to add from the folks at DNAadoption. Adoptees are not just interested in the large segments, the small segments also build the proof of the numerous lines involved. In addition, the accumulation of surnames from all the matches provides a way to evaluate new lines that join into the tree.”
Diane Harman-Hoog (on behalf of the 6 million adoptees in this country, many of who are looking for information on medical records and family heritage).
Diane isn’t the only person who is working with small segment data. Tim Janzen works with small segments, in particular on his Mennonite project, and discusses small segments on the ISOGG WIKI Phasing page. Here is what Tim has to say:
“One advantage of Family Finder is that FF has a 1 cM threshold for matching segments. If a parent and a child both have a matching segment that is in the 2 to 5 cM range and if the number of matching SNPs is 500 or more then there is a reasonably high likelihood that the matching segment is IBD (identical by descent) and not IBS (identical by state).”
The same rules for utilizing larger segment data need to be applied to small segment data to begin with.
Are more guidelines needed for small segments? I don’t know, but we’ll never know if we don’t work with many individual situations and find the common methods for success and identify any problematic areas.
Why Do Small Segments Matter?
In some cases, especially as we work beyond the 6 generation level, small segments may be all we have left of a specific ancestor. If we don’t learn to recognize and utilize the small segments available to us, those ancestors, genetically speaking, will be lost to us forever.
As we move back in time, the DNA from more distant ancestors will be divided into smaller and smaller segments, so if we ever want the ability to identify and track those segments back in time to a specific ancestor, we have to learn how to utilize small segment data – and if we have deleted that data, then we can’t use it.
In my case, I have identified all of my 5th generation ancestors except one, and I have a strong lead on her. In my 6th generation, however, I have lots of walls that need to be broken through – and DNA may be the only way I’ll ever do that.
Let’s take a look at what I can expect when trying to match people who also descend from an ancestor 5 generations back in time. If they are my same generation, they would be my fourth cousins.
Based on the autosomal statistics chart at ISOGG, 4th cousins, on the average, would expect to share about 13.28 cM of DNA from their common ancestor. This would not be over the match threshold at FTDNA of approximately 20 cM total, and if those segments were broken into three pieces, for example, that cousin would not show as a match at either FTDNA or 23andMe, based on the vendors’ respective thresholds.
| % Shared DNA |
Expected Shared cM |
Relationship |
| 0.781% |
53.13 |
Third cousins, common ancestor is 4 generations back in time |
| 0.391% |
26.56 |
Third cousins once removed |
|
20 cm |
Family Tree DNA total cM Threshold |
| 0.195% |
13.28 |
Fourth cousins, common ancestor is 5 generations back in time |
|
7 cM |
23andMe individual segment cM match threshold |
| 0.0977% |
6.64 |
Fourth cousins once removed |
| 0.0488% |
3.32 |
Fifth cousins, common ancestor is 6 generations back in time |
| 0.0244 |
1.66 |
Fifth cousins once removed |
If you’re lucky, as I was with Hickerson, you’ll match at least some relative who carries that ancestral DNA line above the threshold, and then they’ll match other cousins above the threshold, and you can build a comparison network, linking people together, in that fashion. And yes you may well have to utilize GedMatch for people testing at various different vendors and for those smaller segment comparisons.
For clarification, I have never “called” a genealogy match without supporting large segment data. At the vendors, you can’t even see matches if they don’t have larger segments – so there is no way to even know you would match below the threshold.
I do think that we may be able to make calls based on small segments, at least in some instances, in the future. In fact, we have to figure out how to do this or we will rarely be able to move past the 5th or 6th generation utilizing genetics.
At the 5th generation, or third cousins, one expects to see approximately 26 cM of matching DNA, still over the threshold (if divided correctly), but from that point further back in time, the expected shared amount of DNA is under the current day threshold. For those who wonder why the vendors state that autosomal matches are reliable to about the 5th or 6th generation, this is the answer.
I do not discount small segments without cause. In other words, I don’t discount small segments unless there is a reason. Unless they are positively IBS by chance, meaning false, and I can prove it, I don’t disregard them. I do label them and make appropriate notes. You can’t learn from what’s not there.
Let me give you an example. I have one area of my spreadsheet where I have a whole lot of segments, large and small, labeled Acadian. Why? Because the Acadians are so intermarried that I can’t begin to sort out the actual ancestor that DNA came from, at least not yet…so today, I just label them “Acadian.”
This example row is from my master spreadsheet. I have my Mom’s results in my spreadsheet, so I can see easily if someone matches me and Mom both. My rows are pink. The match is on Mom’s side, which I’ve color coded purple. I don’t know which ancestor is the most recent common ancestor, but based on the surnames involved, I know they are Acadian. In some cases, on Acadian matches, I can tell the MRCA and if so, that field is completed as well.

As a note of interest, I inherited my mother’s segment intact, so there was no 50% division in this generation.
I also have segments labeled Mennonite and Brethren. Perhaps in the future I’ll sort through these matches and actually be able to assign DNA segments to specific ancestors. Those segments aren’t useless, they just aren’t yet fully analyzed. As more people test, hopefully, patterns will emerge in many of these DNA groupings, both small and large.
In fact, I talked about DNA patterns and endogamous populations in my recent article, Just One Cousin.
For me, today, some small segment matches appear to be central European matches. I say “appear to be,” because they are not triangulated. For me this is rather boring and nondescript – but if this were my African American client who is trying to figure out which line her European ancestry came from, this could be very important. Maybe she can map these segments to at least a specific ancestral line, which she would find very exciting.
Learning to use small segments effectively has the potential to benefit the following groups of people:
- People with colonial ancestry, because all that may be left today of colonial ancestors is small segments.
- People looking to break down brick walls, not just confirm currently known ancestors.
- People looking for minority ancestors more than 5 or 6 generations back in their trees.
- Adoptees – although very clearly, they want to work with the largest matches first.
- People working with ethnic identification of ancestors, because you will eventually be able to track ethnicity identifying segments back in time to the originating ancestor(s).
Conversely, people from highly endogamous groups may not be helped much, if at all, by small segments because they are so likely to be widely shared within that population as a group from a common ancestor much further back in time. In fact, the definition of a “small segment” for people with fully endogamous families might be much larger than for someone with no known endogamy.
However, if we can identify segments to specific populations, that may help the future accuracy of ethnicity testing.
Let’s go back and take a look at the Hickerson data using the same format we have been using for the comparisons so far.
Small Segment Examples
These Hickerson/Vannoy examples do not utilize random small segment matches, but are utilizing the same matching rules used for larger matches in conjunction with known, triangulated cousin groups from a known ancestor. Many cousins, including 2 brothers and their uncle all carry this same DNA. Like in Israel’s case, where did they get that same DNA if not from a common ancestor?
In the following examples, I want to stress that all of the people involved DO HAVE LARGER SEGMENT MATCHES on other chromosomes, which is how we knew they matched in the first place, so we aren’t trying to prove they are a match. We know they are. Our goal is to determine if small segments are useful in the same situation, proving matches, as with larger segments. In other words, do the rules hold true? And how do we work with the data? Could we utilize these small segment matches if we didn’t have larger matching segments, and if so, how reliable would they be?
There is a difference between a single match and a triangulated group:
- Matches between two people are suggestive of a common ancestor but could be IBS by chance or population..
- Multiple matches, such as with the 6 different Hickersons who descend from Charles Hickerson and Mary Lytle, both in the Ancestry DNA Circle and at Family Tree DNA, are extremely suggestive of a specific common ancestor.
- Only triangulated groups are proof of a common ancestor, unless the people are closely related known relatives.
In our Hickerson/Vannoy study, all participants match at least to one other (but not to all other) group members at Family Tree DNA which means they match over the FTDNA threshold of approximately 20 cM total and at least one segment over 7.7cM and 500 SNPs or more.
In the example below, from the Hickerson article, the known Vannoy cousins are on the left side and the Hickerson matches to the Vannoy cousins are across the top. We have several more now, but this gives you an idea of how the matching stacked up initially. The two green individuals were proven descendants from Charles Hickerson and Mary Lytle.

The goal here is to see how small data segments stack up in a situation where the relationship is distant. Can small segments be utilized to prove triangulation? This is slightly different than in the Just One Cousin article, where the relationship between the individuals was close and previously known. We can contrast the results of that close relationship and small segments with this more distant connection and small segments.
Sarah Hickerson and Daniel Vannoy
The Vannoy project has a group of about a dozen cousins who descend from Elijah Vannoy who have worked together to discover the identify of Elijah’s parents. Elijah’s father is one of 4 Vannoy men, all sons of the same man, found in Wilkes County, NC. in the late 1700s. Elijah Vannoy is 5 generations upstream from me.
What kind of evidence do we have? In the paper genealogy world, I have ruled out one candidate via a Bible record, and probably a second via census and tax records, but we have little information about the third and fourth candidates – in spite of thoroughly perusing all existent records. So, if we’re ever going to solve the mystery, short of that much-wished-for Vannoy Bible showing up on e-Bay, it’s going to have to be via genetic genealogy.
In addition to the dozen or so Vannoy cousins who have DNA tested, we found 6 individuals who descend from Sarah Hickerson’s parents, Charles Hickerson and Mary Lytle who match various Vannoy cousins. Additionally, those cousins match another 21 individuals who carry the Hickerson or derivative surnames, but since we have not proven their Hickerson lineage on paper, I have not utilized any of those additional matches in this analysis. Of those 26 total matches, at Family Tree DNA, one Hickerson individual matches 3 Vannoy cousins, nine Hickerson descendants match 2 Vannoy cousins and sixteen Hickerson descendants match 1 Vannoy cousin.
Our group of Vannoy cousins matching to the 6 Charles Hickerson/Mary Lytle descendants contains over 60 different clusters of matching DNA data across the 22 chromosomes. Those 6 individuals are included in 43 different triangulated groups, proving the entire triangulation group shares a common ancestor. And that is BEFORE we add any GedMatch information.
If that sounds like a lot, it’s not. Another recent article found 31 clusters among siblings and their first cousin, so 60 clusters among a dozen known Vannoy cousins and half a dozen potential Hickerson cousins isn’t unusual at all.
To be very clear, Sarah Hickerson and Daniel Vannoy were not “declared” to be the parents of Elijah Vannoy, born in 1784, based on small segment matches alone. Larger segment matches were involved, which is how we saw the matches in the first place. Furthermore, the matches triangulated. However, small segments certainly are involved and are more prevalent, of course, than large segments. Some cousins are only connected by small segments. Are they valid, and how do we tell? Sometimes it’s all we have.
Let me give you the classic example of when small segments are needed.
We have four people. Person A and B are known Vannoy cousins and person C and D are potential Hickerson cousins. Potential means, in this case, potential cousins to the Vannoys. The Hickersons already know they both descend from Charles Hickerson and Mary Lytle.
- Person A matches person C on chromosome 1 over the matching threshold.
- Person B matches person D on chromosome 2 over the matching threshold.
Both Vannoy cousins match Hickerson cousins, but not the same cousin and not on the same segments at the vendor. If these were same segment matches, there would be no question because they would be triangulated, but they aren’t.
So, what do we do? We don’t have access to see if person C and D match each other, and even if we did, they don’t match on the same segments where they match persons A and B, because if they did we’d see them as a match too when we view A and B.
If person A and B don’t match each other at the vendor, we’re flat out of luck and have to move this entire operation to GedMatch, assuming all 4 people have or are willing to download their data.

If person A and B match each other at the vendor, we can see their small segment data as compared to each other and to persons C and D, respectively which then gives us the ability to see if A matches C on the same small segment as B matches D.

If we are lucky, they will all show a common match on a small segment – meaning that A will match B on a small segment of chromosome 3, for example, and A will match C on that same segment. In a perfect world, B will also match D on that same segment, and you will have 4 way triangulation – but I’m happy with the required 3 way match to triangulate.
This is exactly what happened in the article, Be Still My H(e)art. As you can see, three people match on chromosomes 1 and 8, below – two of whom are proven cousins and the third was the wife surname candidate line.
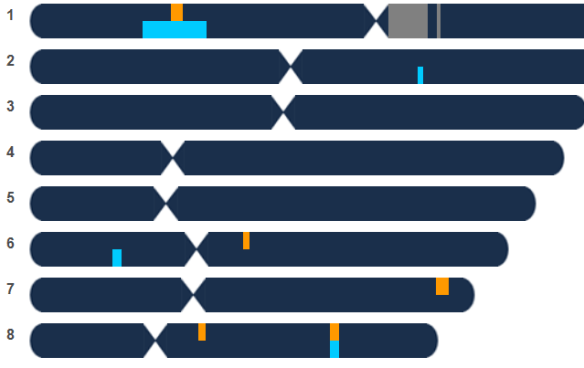
The example I showed of chromosome 2 in the Hickerson article was where all participants of the 5 individuals shown on the chromosome browser were matching to the Vannoy participant. I thought it was a good visual example. It was just one example of the 60+ clusters of cousin matches between the dozen Vannoy cousins and 6 Hickerson descendants.
This example was criticized by some because it was a small segment match. I should probably have utilized chromosome 15 or searched for a better long segment example, but the point in my article was only to show how people that match stack up together on the chromosome browser – nothing more. Here’s the entire chromosome, for clarity.

Certainly, I don’t want to mislead anyone, including myself. Furthermore, I dislike being publicly characterized as “wrong” and worse yet, labeled “irresponsible,” so I decided to delve into the depths of the data and work through several different examples to see if small segment data matching holds in various situations. Let’s see what we found.
Chromosome 15
I selected chromosome 15 to work with because it is a region where a lot of Vannoy descendants match – and because it is a relatively large segment. If the Hickersons do match the Vannoys, there’s a fairly good change they might match on at least part of that segment. In other words, it appears to be my best bet due to sheer size and the number of Elijah Vannoy’s descendants who carry this segment. In addition to the 6 individuals above who matched on chromosome 15, here are an additional 4. As you can see, chromosome 15 has a lot of potential.

The spreadsheet below shows the sections of chromosome 15 where cousins match. Green individuals in the Match column are descendants of Charles Hickerson and Mary Lytle, the parents of Sarah Hickerson. The balance are Vannoys who match on chromosome 15.

As you can see, there are several segments that are quite large, shown in yellow, but there are also many that are under the threshold of 7cM, which are all segments that would be deleted if you are deleting small segments. Please also note that if you were deleting small segments, all of the Hickerson matches would be gone from chromosome 15.
Those of you with an eagle eye will already notice that we have two separate segments that have triangulated between the Vannoy cousins and the Hickerson descendants, noted in the left column by yellow and beige. So really, we could stop right here, because we’ve proven the relationship, but there’s a lot more to learn, so let’s go on.
You Can’t Use What You Can’t See
I need to point something out at this point that is extremely important.
The only reason we see any segment data below the match threshold is because once you match someone on a larger segment at Family Tree DNA, over the threshold, you also get to view the small segment data down to 1cM for your match with that person.
What this means is that if one person or two people match a Hickerson descendant, for example you will see the small segment data for their individual matches, but not for anyone that doesn’t match the participant over the matching threshold.
What that means in the spreadsheet above, is that the only Hickerson that matches more than one Vannoy (on this segment) is Barbara – so we can see her segment data (down to 1cM ) as compared to Polly and Buster, but not to anyone else.
If we could see the smaller segment data of the other participants as compared to the Hickerson participants, even though they don’t match on a larger segment over the matching threshold, there could potentially be a lot of small segment data that would match – and therefore triangulate on this segment.
This is the perfect example of why I’ve suggested to Family Tree DNA that within projects or in individuals situations, that we be allowed to reduce the match threshold – especially when a specific family line match is suspected.
This is also one of the reasons why people turn to GedMatch, and we’ll do that as well.
What this means, relative to the spreadsheet is that it is, unfortunately, woefully incomplete – and it’s not apples to apples because in some cases we have data under the match threshold, and in some, we don’t. So, matches DO count, but nonmatches where small segment data is not available do NOT count as a non-match, or as disproof. It’s only negative proof IF you have the data AND it doesn’t match.
The Vannoys match and triangulate on many segments, so those are irrelevant to this discussion other than when they match to Hickerson DNA. William (H), descends from two sons of Charles Hickerson and Mary Lytle. Unfortunately, he only matches one Vannoy, so we can only see his small segments for that one Vannoy individual, William (V). We don’t know what we are missing as compared to the rest of the Vannoy cousins.
To see William (H)’s and William (V)’s DNA as compared to the rest of the Vannoy cousins, we had to move to GedMatch.
Matching Options
Since we are working with segments that are proven to be Vannoy, and we are trying to prove/disprove if Daniel Vannoy and Sarah Hickerson are the parents of Elijah through multiple Hickerson matches, there are only a few matching options, which are:
- The Hickerson individuals will not triangulate with any of the Vannoy DNA, on chromosome 15 or on other chromosomes, meaning that Sarah Hickerson is probably not the mother of Elijah Vannoy, or the common ancestor is too far back in time to discern that match at vendor thresholds.
- The Hickerson individuals will not triangulate on this segment, but do triangulate on other segments, meaning that this segment came entirely from the Vannoy side of the family and not the Hickerson side of the family. Therefore, if chromosome 15 does not triangulate, we need to look at other chromosomes.
- The Hickerson individuals triangulate with the Vannoy individuals, confirming that Sarah Hickerson is the mother of Elijah Vannoy, or that there is a different common unknown ancestor someplace upstream of several Hickersons and Vannoys.
All of the Vannoy cousins descend from Elijah Vannoy and Lois McNiel, except one, William (V), who descends from the proven son of Sarah Hickerson and Daniel Vannoy, so he would be expected to match at least some Hickerson descendants. The 6 Hickerson cousins descend from Charles Hickerson and Mary Lytle, Sarah’s parents.

William (H), the Hickerson cousin who descends from David, brother to Sarah Hickerson, is descended through two of David Hickerson’s sons.
I decided to utilize the same segment “mapping comparison” technique with a spreadsheet that I utilized in the phasing article, because it’s easy to see and visualize.
I have created a matching spreadsheet and labeled the locations on the spreadsheet from 25-100 based on the beginning of the start location of the cluster of matches and the end location of the cluster.
Each individual being compared on the spreadsheet below has a column across the top. On the chart below, all Hickerson individuals are to the right and are shown with their cells highlighted yellow in the top row.
Below, the entire colorized chart of chromosome 15 is shown, beginning with location 25 and ending with 100, in the left hand column, the area of the Vannoy overlap. Remember, you can double click on the graphics to enlarge. The columns in this spreadsheet are not fully expanded below, but they are in the individual examples.

I am going to step through this spreadsheet, and point out several aspects.
First, I selected Buster, the individual in the group to begin the comparison, because he was one of the closest to the common ancestor, Elijah Vannoy, genealogically, at 4 generations. So he is the person at Family Tree DNA that everyone is initially compared against.
Everyone who matches Buster has their matching segments shown in blue. Buster is shown furthest left.
When participants match someone other than Buster, who they match on that segment is typed into their column. You can tell who Buster matches because their columns are blue on matching locations. Here’s an example.

You can see that in my column, it’s blue on all segments which means I match Buster on this entire region. In addition, there are names of Carl, Dean, William Gedmatch and Billie Gedmatch typed into the cell in the first row which means at that location, in addition to Buster, I also match Carl and Dean at Family Tree DNA and William (descended from the son of Daniel Vannoy and Sarah Hickerson) at Gedmatch and Billie (a Hickerson) at Gedmatch. Their name is typed into my column, and mine into theirs. Please note that I did not run everyone against everyone at GedMatch. I only needed enough data to prove the point and running many comparisons is a long, arduous process even when GedMatch isn’t experiencing problems.
On cells that aren’t colorized blue, the person doesn’t match Buster, but may still match other Vannoy cousin segments. For example, Dean, below, matches Buster on location 25-29, along with some other cousins. However, he does not match Buster on location 30 where he instead matches Harold and Carl who also don’t match Buster at that location. Harold, Carl and Dean do, however, all descend from the same son of Elijah so they may well be sharing DNA from a Vannoy wife at this location, especially since no one who doesn’t share that specific wife’s line matches those three at this location.

Remember, we are not working with random small data segments, but with a proven matching segment to a common Vannoy ancestor, with a group of descendants from a possible/probable Hickerson ancestor that we are trying to prove/disprove. In other words, you would expect either a lot of Hickerson matches on the same segments, if Hickerson is indeed a Vannoy ancestral family, or virtually none of them to match, if not.
The next thing I’d like to point out is that these are small segments of people who also have larger matching segments, many of whom do triangulate on larger segments on other chromosomes. What we are trying to discern is whether small segment matches can be utilized by employing the same matching criteria as large segment matching. In other words, is small segment data valid and useful if it meets the criteria for an IBD match?
For example, let’s look at Daniel. Daniel’s segments on chromosome 15, were it not for the fact that he matches on larger segments on other chromosomes, would not be shown as matches, because they are not individually over the match threshold.
Look at Daniel’s column for Polly and Warren.

The segments in red show a triangulated group where Daniel and Warren, or Daniel, Warren and Polly match. The segments where all 3 match are triangulated.
This proves, unquestionably, that small segments DO match utilizing the normal prescribed IBD matching criteria. This spreadsheet, just for chromosome 15, is full of these examples.
Is there any reason to think that these triangulated matches are not identical by descent? If they are not IBD, how do all of these people match the same DNA? Chance alone? How would that be possible? Two people, yes, maybe, but 3 or more? In some cases, 5 or 6 on the same segment? That is simply not possible, or we have disproven the entire foundation that autosomal DNA matching is based upon.
The question will soon be asked if small segments that triangulate can be useful when there are no larger matching segments to put the match over the initial vendor threshold.
Triangulated Groups
As you can see, most of the people and segments on the spreadsheet, certainly the Elijah descendants, are heavily triangulated, meaning that three or more people match each other on the same locations. Most of this matching is over the vendor threshold at Family Tree DNA.
You can see that Buster, Me, Dean, Carl and Harold all match each other on the same segments, on the left half of the spreadsheet where our names are in each other’s columns.

Remember when I said that the spreadsheet was incomplete? This is an example. David and Warren don’t match each other at a high enough total of segments to get them over the matching threshold when compared to each other, so we can’t see their small segment data as compared to each other. David matches Buster, but Warren doesn’t, so I can’t even see them both in relationship to a common match. There are several people who fall into this category.
Let’s select one individual to use as an example.
I’ve chosen the Vannoy cousin, William(V), because his kit has been uploaded to Gedmatch, he has Vannoy matches and because William is proven to descend from Sarah Hickerson and Daniel Vannoy through their son Joel – so we expect some Hickerson DNA to match William(V).
If William (V) matches the Hickersons on the same DNA locations as he matches to Elijah’s descendants, then that proves that Elijah’s descendant’s DNA in that location is Hickerson DNA.
At GedMatch, I compared William(V) with me and then with Dean using a “one to one” comparison at a low threshold, simply because I wanted as much data as I could get. Family Tree DNA allows for 1 cM and I did the same, allowing 100 SNPs at GedMatch. Family Tree DNA’s lowest SNP threshold is 500.
In case you were wondering, even though I did lower the GedMatch threshold below the FTDNA minimum, there were 45 segments that were above 1cM and above 500 SNPs when matching me to William(V), which would have been above the lowest match threshold at FTDNA (assuming we were over the initial match threshold.) In other words, had we not been below the original match threshold (20cM total, one segment over 7.7cM), these segments would have been included at FTDNA as small segments. As you can see in the chart below, many triangulated.
I colorized the GedMatch matches, where there were no FTDNA matches, in dark red text. This illustrates graphically just how much is missed when the small segments are ignored in cases with known or probable cousins. In the green area, the entry that says “Me GedMatch” could not be colorized red (because you can’t colorize only part of the text of a cell) so I added the Gedmatch designation to differentiate between a match through FTDNA and one from GedMatch. I did the same with all Gedmatch matches, whether colorized or not.
Let’s take a look and see how small segments from GedMatch affect our Hickerson matching. Note that in the green area, William (V) matches William (H), the Hickerson descendant, and William (V) matches to me and Dean as well. This triangulates William (V)’s Hickerson DNA and proves that Elijah’s descendants DNA includes proven Hickerson segments.

In this next example, I matched William (H), the Hickerson cousin (with no Vannoy heritage) against both Buster and me.

Without Gedmatch data, only two segments of chromosome 15 are triangulated between Vannoy and Hickerson cousins, because we can’t see the small data segments of the rest of the cousins who don’t match over the threshold.
You can see here that nearly the entire chromosome is triangulated using small segments. In the chart below, you can see both William(V) and William (H) as they match various Vannoy cousins. Both triangulate with me.

I did the same thing with the Hickerson descendant, Billie, as compared to both me and Dean, with the same type of results.
The next question would be if chromosome 15 is a pileup area where I have a lot of IBS matches that are really population based matches. It does not appear to be. I have identified an area of my chromosomes that may be a pileup area, but chromosome 15 does not carry any of those characteristics.
So by utilizing the small segments at GedMatch for chromosome 15 that we can’t otherwise see, we can triangulate at least some of the Hickerson matches. I can’t complete this chart, because several individuals have not uploaded to GedMatch.
Why would the Hickerson descendant match so many of the Vannoy segments on chromosome 15? Because this is not a random sample. This is a proven Vannoy segment and we are trying to see which parts of this segment are from a potential Hickerson mother or the Vannoy father. If from the Hickerson mother, then this level of matching is not unexpected. In fact, it would be expected. Since we cheated and saw that chromosome 15 was already triangulated at Family Tree DNA, we already knew what to expect.
In the spreadsheet below, I’ve added the 2 GedMatch comparisons, William (V) to me and Dean, and William (H) to me and Buster. You can see the segments that triangulate, on the left. We could also build “triangulated groups,” like GedMatch does. I started to do this, but then stopped because I realized most cells would be colored and you’d have a hard time seeing the individual triangulated segments. I shifted to triangulating only the individuals who triangulate directly with the Hickerson descendant, William(H), shown in green. GedMatch data is shown in red.

I would like to make three points.
1. This still is not a complete spreadsheet where everyone is compared to everyone. This was selectively compared for two known Hickerson cousins, William (V) who descends from both Vannoys and Hickersos and William (H) who descends only from Hickersons.
2. There are 25 individually triangulated segments to the Hickerson descendant on just this chromosome to the various Vannoy cousins. That’s proof times 25 to just one Hickerson cousin.
3. I would NEVER suggest that you select one set of small segments and base a decision on that alone. This entire exercise has assembled cumulative evidence. By the same token, if the rules for segment matching hold up under the worst circumstances, where we have an unknown but suspected relationship and the small segments appear to continue to follow the triangulation rules, they could be expected to remain true in much more favorable circumstances.
Might any of these people have random DNA matches that are truly IBS by chance on chromosome 15? Of course, but the matching rules, just like for larger segments, eliminates them. According to triangulation rules, if they are IBS by chance, they won’t triangulate. If they do triangulate, that would confirm that they received the same DNA from a common ancestor.
If this is not true, and they did not receive their common DNA from a common ancestor, then it disproves the fundamental matching rule upon which all autosomal DNA genetic genealogy is based and we all need to throw in the towel and just go and do something else.
Is there some grey area someplace? I would presume so, but at this point, I don’t know how to discern or define it, if there is. I’ve done three in-depth studies on three different families over the past 6 weeks or so, and I’ve yet to find an area (except for endogamous populations that have matches by population) where the guidelines are problematic. Other researchers may certainly make different discoveries as they do the same kind of studies. There is always more to be discovered, so we need to keep an open mind.
In this situation, it helps a lot that the Hickerson/Vannoy descendants match and triangulate on larger segments on other chromosomes. This study was specifically to see if smaller segments would triangulate and obey the rules. We were fortunate to have such a large, apparently “sticky” segment of Vannoy DNA on chromosome 15 to work with.
Does small segment matching matter in most cases, especially when you have larger segments to utilize? Probably not. Use the largest segments first. But in some cases, like where you are trying to prove an ancestor who was born in the 1700s, you may desperately need that small segment data in order to triangulate between three people.
Why is this important – critically important? Because if small segments obey all of the triangulation rules when larger segments are available to “prove” the match, then there is no reason that they couldn’t be utilized, using the same rules of IBD/IBS, when larger segments are not available. We saw this in Just One Cousin as well.
However, in terms of proof of concept, I don’t know what better proof could possibly be offered, within the standard genetic genealogy proofs where IBD/IBS guidelines are utilized as described in the Phasing article. Additional examples of small segment proof by triangulation are offered in Just One Cousin, Lazarus – Putting Humpty Dumpty Together Again, and in Demystifying Autosomal DNA Matching.
Raising Elijah Vannoy and Sarah Hickerson from the Dead
As I thought more about this situation, I realized that I was doing an awful lot of spreadsheet heavy lifting when a tool might already be available. In fact, Israel’s mention of Lazarus made me wonder if there was a way to apply this tool to the situation at hand.
I decided to take a look at the Lazarus tool and here is what the intro said:
Generate ‘pseudo-DNA kits’ based on segments in common with your matches. These ‘pseudo-DNA kits’ can then be used as a surrogate for a common ancestor in other tests on this site. Segments are included for every combination where a match occurs between a kit in group1 and group2.
It’s obvious from further instructions that this is really meant for a parent or grandparent, but the technique should work just the same for more distant relatives.
I decided to try it first just with the descendants of Elijah Vannoy. At first, I thought that recreated Elijah would include the following DNA:
- DNA segments from Elijah Vannoy
- DNA segments from Elijah Vannoy’s wife, Lois McNiel
- DNA segments that match from Elijah’s descendants spouse’s lines when individuals come from the same descendant line. This means that if three people descend from Joel Vannoy and Phoebe Crumley, Elijah’s son and his wife, that they would match on some DNA from Phoebe, and that there was no way to subtract Phoebe’s DNA.
After working with the Lazarus tool, I realized this is not the case because Lazarus is designed to utilize a group of direct descendants and then compare the DNA of that group to a second group of know relatives, but not descendants.
In other words, if you have a grandson of a man, and his brother. The DNA shared by the brother and the grandson HAS to be the DNA contributed to that grandson by his grandfather, from their common ancestor, the great grandfather. So, in our situation above, Phoebe’s DNA is excluded.
The chart below shows the inheritance path for Lazarus matching.

Because Lazarus is comparing the DNA of Son Doe with Brother Doe – that eliminates any DNA from the brother’s wives, Sarah Spoon or Mary – because those lines are not shared between Brother Doe and Son Doe. The only shared ancestors that can contribute DNA to both are Father Doe and Methusaleh Fisher.
The Lazarus instructions allow you to enter the direct descendants of the person/couple that you are reconstructing, then a second set of instructions asks for remaining relatives not directly descended, like siblings, parents, cousins, etc. In other words, those that should share DNA through the common ancestor of the person you are recreating.
To recreate Elijah, I entered all of the Vannoy cousins and then entered William (V) as a sibling since he is the proven son of Daniel Vannoy and Sarah Hickerson.
Here is what Lazarus produced.

Lazarus includes segments of 4cM and 500 SNPs.
The first thing I thought was, “Holy Moly, what happened to chromosome 15?” I went back and looked, and sure enough, while almost all of the Elijah descendants do match on chromosome 15, William (V), kit 156020, does not match above the Lazarus threshold I selected. So chromosome 15 is not included. Finding additional people who are known to be from this Vannoy line and adding them to the “nondescendant” group would probably result in a more complete Elijah.

Next, to recreate Sarah Hickerson, I added all of the Vannoy cousins plus William (V) as descendants of Sarah Hickerson and then I added just the one Hickerson descendant, William, as a sibling. William’s ancestor is proven to be the sibling of Sarah.
I didn’t know quite what to expect.
Clearly if the DNA from the Hickerson descendant didn’t match or triangulate with DNA from any of the Vannoy cousins at this higher level, then Sarah Hickerson wasn’t likely Elijah’s mother. I wanted to see matching, but more, I wanted to see triangulation.

I was stunned. Every kit except two had matches, some of significant size.


Please note that locations on chromosomes 3, 4 and 13, above, are triangulated in addition to matching between two individuals, which constitutes proof of a common ancestor. Please also note that if you were throwing away segments below 7cM, you would lose all of the triangulated matches and all but two matches altogether.
Clearly, comparing the Vannoy DNA with the Hickerson DNA produced a significant number of matches including three triangulated segments.

Where Are We?
I never have, and I never would recommend attempting to utilize random small match segments out of context. By out of context, I mean simply looking at all of your 1cM segments and suggesting that they are all relevant to your genealogy. Nope, never have. Never would.
There is no question that many small segments are IBS by chance or identical by population. Furthermore, working with small segments in endogamous populations may not be fruitful.
Those are the caveats. Small segments in the right circumstances are useful. And we’ve seen several examples of the right circumstances.
Over the past few weeks, we have identified guidelines and tools to work with small segments, and they are the same tools and guidelines we utilize to work with larger segments as well. The difference is size. When working with large segments, the fact that they are large serves an a filter for us and we don’t question their authenticity. With all small segments, we must do the matching and analysis work to prove validity. Probably not worthwhile if you have larger segments for the same group of people.
Working with the Vannoy data on chromosome 15 is not random, nor is the family from an endogamous population. That segment was proven to be Vannoy prior to attempts to confirm or disprove the Hickerson connection. And we’ve gone beyond just matching, we’ve proven the ancestral link by triangulation, including small segments. We’ve now proven the Hickerson connection about 7 ways to Sunday. Ok, maybe 7 is an exaggeration, but here is the evidence summed up for the Vannoy/Hickerson study from multiple vendors and tools:
- Ancestry DNA Circle indicating that multiple Hickerson descendants match me and some that don’t match me, match each other. Not proof, but certainly suggestive of a common ancestor.
- A total of 26 Hickerson or derivative family name matches to Vannoy cousins at Family Tree DNA. Not proof, but again, very suggestive.
- 6 Charles Hickerson/Mary Lytle descendants match to Vannoy cousins at Family Tree DNA. Extremely suggestive, needs triangulation.
- Triangulation of segments between Vannoy and Hickerson cousins at Family Tree DNA. Proof, but in this study we were only looking to determine whether small segment matches constituted proof.
- Triangulation of multiple Hickerson/Vannoy cousins on chromosome 15 at GedMatch utilizing small segments and one to one matching. More proof.
- Lazarus, at higher thresholds than the triangulation matching, when creating Sarah Hickerson, still matched 19 segments and triangulated three for a total of 73.2cM when comparing the Hickerson descendant against the Vannoy cousins. Further proof.
So, can small segment matching data be useful? Is there any reason NOT to accept this evidence as valid?
With proper usage, small segment data certainly looks to provide value by judiciously applying exactly the same rules that apply to all DNA matching. The difference of course being that you don’t really have to think about utilizing those tools with large segment matches. It’s pretty well a given that a 20cM match is valid, but you can never assume anything about those small segment matches without supporting evidence. So are larger segments easier to use? Absolutely.
Does that automatically make small segments invalid? Absolutely not.
In some cases, especially when attempting to break down brick walls more than 5 or 6 generations in the past, small segment data may be all we have available. We must use it effectively. How small is too small? I don’t know. It appears that size is really not a factor if you strictly adhere to the IBD/IBS guidelines, but at some point, I would think the segments would be so small that just about everyone would match everyone because we are all humans – so the ultimate identical by population scenario.
Segments that don’t match an individual and either or both parents, assuming you have both parents to test, can safely be disregarded unless they are large and then a look at the raw data is in order to see if there is a problem in that area. These are IBS by chance. IBS segments by chance also won’t triangulate further up the tree. They can’t, because they don’t match your parents so they cannot come from an ancestor. If they don’t come from an ancestor, they can’t possibly match two other people whose DNA comes from that ancestor on that segment.
If both parents aren’t available, or your small segments do match with your parents, I would suggest that you retain your small segments and map them.
You can’t recognize patterns if the data isn’t present and you won’t be able to find that proverbial needle in the haystack that we are all looking for.
Based on what we’ve seen in multiple case studies, I would conclude that small segment data is certainly valid and can play a valid role in a situation where there is a known or suspected relationship.
I would agree that attempting to utilize small segment data outside the context of a larger data match is not optimal, at least not today, although I wish the vendors would provide a way for us to selectively lower our thresholds. A larger segment match can point the way to smaller segment matches between multiple people that can be triangulated. In some situations, like the person A, B, C, D Hickerson-Vannoy situation I described earlier in this article, I would like to be able to drop the match threshold to reveal the small segment data when other matches are suggestive of a family relationship.
In the Hickerson situation, having the ability to drop the matching thresholds would have been the key to positively confirming this relationship within the vendor’s data base and not having to utilize third party tools like GedMatch – which require the cooperation of all parties involved to download their raw data files. Not everyone transferred their data to Gedmatch in my Vannoy group, but enough did that we were able to do what we needed to do. That isn’t always the case. In fact, I have an nearly identical situation in another line but my two matches at Ancestry have declined to download their data to Gedmatch.
This not the first time that small segment data has played a successful role in finding genealogy solutions, or confirming what we thought we knew – although in all cases to date, larger segments matched as well – and those larger segment matches were key and what pointed me to the potential match that ultimately involved the usage of the small segments for triangulation.
Using larger data segments as pointers probably won’t be the case forever, especially if we can gain confidence that we can reliably utilize small segments, at least in certain situations. Specifically, a small segment match may be nothing, but a small segment triangulated match in the context of a genealogical situation seems to abide by all of the genetic genealogy DNA rules.
In fact, a situation just arose in the past couple weeks that does not include larger segments matching at a vendor.
Let’s close this article by discussing this recent scenario.
The Adoptee
An adoptee approached me with matching data from GedMatch which included matches to me, Dean, Carl and Harold on chromosome 15, on segments that overlap, as follows.

On the spreadsheet above, sent to me by the adoptee, we can see some matches but not all matches. I ran the balance of these 4 people at GedMatch and below is the matching chart for the segment of chromosome 15 where the adoptee matches the 4 Vannoy cousins plus William(H), the Hickerson cousin.
| |
Me |
Carl |
Dean |
Harold |
Adoptee |
| Me |
NA |
FTDNA |
FTDNA |
GedMatch |
GedMatch |
| Carl |
FTDNA |
NA |
FTDNA |
FTDNA |
GedMatch |
| Dean |
FTDNA |
FTDNA |
NA |
FTDNA |
GedMatch |
| Harold |
GedMatch |
FTDNA |
FTDNA |
NA |
GedMatch |
| Adoptee |
GedMatch |
GedMatch |
GedMatch |
GedMatch |
NA |
| William (H) |
GedMatch |
GedMatch |
GedMatch |
GedMatch |
GedMatch |
I decided to take the easy route and just utilize Lazarus again, so I added all of the known Vannoy and Hickerson cousins I utilized in earlier Lazarus calculations at Gedmatch as siblings to our adoptee. This means that each kit will be compared to the adoptees DNA and matching segments will be reported. At a threshold of 300 SNPs and 4cM, our adoptee matches at 140cM of common DNA between the various cousins.

Please note that in addition to matching several of the cousins, our adoptee also triangulates on chromosomes 1, 11, 15, 18, 19 and 21. The triangulation on chromosome 21 is to two proven Hickerson descendants, so he matches on this line as well.
I reduced the threshold to 4cM and 200 SNPs to see what kind of difference that would make.

Our adoptee picked up another triangulation on chromosome 1 and added additional cousins in the chromosome 15 “sticky Vannoy” cluster and the chromosome 18 cluster.
Given what we just showed about chromosome 15, and the discussions about IBD and IBS guidelines and small matching segments, what conclusions would you draw and what would you do?
- Tell the adoptee this is invalid because there are no qualifying large match segments that match at the vendors.
- Tell the adoptee to throw all of those small segments away, or at least all of the ones below 7cM because they are only small matching segments and utilizing small matching segments is only a folly and the adoptee is only seeing what he wants to see – even though the Vannoy cousins with whom he triangulates are proven, triangulated cousins.
- Check to see if the adoptee also matches the other cousins involved, although he does clearly already exceeds the triangulation criteria to declare a common ancestor of 3 proven cousins on a matching segment. This is actually what I did utilizing Lazarus and you just saw the outcome.
If this is a valid match, based on who he does and doesn’t match in terms of the rest of the family, you could very well narrow his line substantially – perhaps by utilizing the various Vannoy wives’ DNA, to an ancestral couple. Given that our adoptee matches both the Vannoys and the Hickersons, I suspect he is somehow descended from Daniel Vannoy and Sarah Hickerson.
In Conclusion
What is the acceptable level to utilize small segments in a known or suspected match situation?
Rather than look for a magic threshold number, we are much better served to look at reliable methods to determine the difference between DNA passed from our ancestors to us, IBD, and matches by chance. This helps us to establish the reliability of DNA segments in individual situations we are likely to encounter in our genealogy. In other words, rather that throw the entire pile of wheat away because there is some percentage of chaff in the wheat, let’s figure out how to sort the wheat from the chaff.
Fortunately, both parental phasing and triangulation eliminate the identical by chance segments.
Clearly, the smaller the segments, even in a known match situation, the more likely they are identical by population, given that they triangulate. In fact, this is exactly how the Neanderthal and Denisovan genomes have been reconstructed.
Furthermore, given that the Anzick DNA sample is over 12,000 years old, Identical by population must be how Anzick is matching to contemporary humans, because at least some of these people do clearly share a common ancestor with Anzick at some point, long ago – more than 12,000 years ago. In my case, at least some of the Anzick segments triangulate with my mother’s DNA, so they are not IBS by chance. That only leaves identical by population or identical by descent, meaning within a genealogical timeframe, and we know that isn’t possible.
There are yet other situations where small segment matches are not IBS by chance nor identical by population. For example, I have a very hard time believing that the adoptee situation is nothing but chance. It’s not a folly. It’s identical by descent as proven by triangulation with 10 different cousins – all on segments below the vendor matching thresholds.
In fact, it’s impossible to match the Vannoy cousins, who are already triangulated individually, by chance. While the adoptee match is not over the vendor threshold, the segments are not terribly small and they do all triangulate with multiple individuals who also triangulate with larger segments, at the vendors and on different chromosomes.
This adoptee triangulated match, even without the Hickerson-Vannoy study disproves the blanket statement that small segments below 5cM cannot be used for genealogy. All of these segments are 7.1cM or below and most are below 5.
This small segment match between my mother and her first cousins also disproves that segments under 5cM can never be used for genealogy.

This small segment passed from my mother to me disproves that statement too – clearly matching with our cousin, Cheryl. If I did not receive this from my mother, and she from her parent, then how do we match a common cousin???

More small segment proof, below, between my mother and her second cousin when Lazarus was reconstructing my mother’s father.

And this Vannoy Hickerson 4 cousin triangulated segment also disproves that 5cM and below cannot be used for genealogy.

Where did these small segments come from if not a common ancestor, either one or several generations ago? If you look at the small segment I inherited from my mother and say, “well, of course that’s valid, you got it from your mother” then the same logic has to apply that she inherited it from her parent. The same logic then applies that the same small segment, when shared by my mother’s cousin, also came from the their common grandparents. One cannot be true without the others being true. It’s the same DNA. I got it from my mother. And it’s only a 1.46cM segment, shown in the examples above.
Here are my observations and conclusions:
- As proven with hundreds of examples in this and other articles cited, small segments can be and are inherited from our ancestors and can be utilized for genetic genealogy.
- There is no line in the sand at 7cM or 5cM at which a segment is viable and useful at 5.1cM and not at 4.9cM.
- All small segment matches need to be evaluated utilizing the guidelines set forth for IBD versus IBS by chance versus identical by population set forth in the articles titled How Phasing Works and Determining IBD Versus IBS Matches and Demystifying Autosomal DNA Matching.
- When given a choice, large segment matches are always easier to use because they are seldom IBS by chance and most often IBD.
- Small segment matches are more likely to be IBS by chance than larger matches, which is why we need to judiciously apply the IBD/IBS Guidelines when attempting to utilize small segment matches.
- All DNA matches, not just small segments, must be triangulated to prove a common ancestor, unless they are known close relatives, like siblings, first cousins, etc.
- When working in genetic genealogy, always glean the information from larger matches and assemble that information. However, when the time comes that you need those small segments because you are working 5, 6 or 7 generations back in time, remember that tools and guidelines exist to use small segments reliably.
- Do not attempt to use small segments out of context. This means that if you were to look only at your 1cM matches to unknown people, and you have the ability to triangulate against your parents, most would prove to be IBS by chance. This is the basis of the argument for why some people delete their small segments. However, by utilizing parental phasing, phasing against known family members (like uncles, aunts and first cousins) and triangulation, you can identify and salvage the useable small segments – and these segments may be the only remnants of your ancestors more than 5 or 6 generations back that you’ll ever have to work with. You do not have to throw all of them away simply because some or many small segments, out of context, are IBS by chance. It doesn’t hurt anything to leave them just sit in your spreadsheet untouched until the day that you need them.
Ultimately, the decision is yours whether you will use small segments or not – and either decision is fine. However, don’t make the decision based on the belief that small segments under some magic number, like 5cM or 7cM are universally useless. They aren’t.
Whether small segments are too much work and effort in your individual situation depends on your personal goals for genetic genealogy and on factors like whether or not you descend from an endogamous population. People’s individual goals and circumstances vary widely. Some people test at Ancestry and are happy with inferential matching circles and nothing more. Some people want to wring every tidbit possible out of genealogy, genetic or otherwise.
I hope everyone will begin to look at how they can use small segment data reliably instead of simply discarding all the small segments on the premise that all small segment data is useless because some small segments are not useful. All unstudied and discarded data is indeed useless, so discarding becomes a self-fulfilling prophecy.
But by far, the worst outcome of throwing perfectly good data away is that you’ll never know what genetic secrets it held for you about your ancestors. Maybe the DNA of your own Sarah Hickerson is lurking there, just waiting for the right circumstances to be found.
______________________________________________________________
Disclosure
I receive a small contribution when you click on some of the links to vendors in my articles. This does NOT increase the price you pay but helps me to keep the lights on and this informational blog free for everyone. Please click on the links in the articles or to the vendors below if you are purchasing products or DNA testing.
Thank you so much.
DNA Purchases and Free Transfers
Genealogy Services
Genealogy Research
Like this:
Like Loading...