I didn’t want to open the curtains Saturday morning, given the blizzard warnings that were worsening all day Friday.
I finally screwed up my courage and peeked out.
Indeed, those skies look dark, grey, and foreboding.
Decision Time
I had a decision to make.
I originally planned to stay in Salt Lake City until Sunday, but I had already changed my flight to late Saturday afternoon following my session. I also changed my hotel reservation accordingly.
However, if I packed and checked out, got to the airport, and my flight was canceled, I was likely going to be stranded, potentially at the airport for at least two days. There aren’t any hotels in the Salt Lake City airport, but there are a few nearby. However, probably not enough rooms to accommodate an airport full of stranded people.
Would Uber even be available?
Could I get back into Salt Lake City to check back into the Marriott or any other hotel? Would they have space?
I was at the go-no-go decision point.
It was probably a 50-50 roll of the dice.
I packed and checked out.
It wasn’t snowing yet, but no one doubted that it would. The only questions were when the snow would begin, whether it would begin as rain and freeze into ice before the snow started falling, and how much snow would there be.
Maybe more important to the people at RootsTech – what about flights?
All day, you could see people obsessively checking their flight information on their phones.
I had two speaking engagements scheduled for the day: a morning Y-DNA “Ask Me Anything” panel and my afternoon session, “Highways of History – Flesh out Your Ancestors Using Discover Case Studies.”
The afternoon session was scheduled to end just half an hour before RootsTech closed for the year, so there really was no getting out early.
In for a penny, in for a pound.
Friends in the Expo Hall
I was still trying to visit every booth on the show floor.
I’ll just admit right now that I failed miserably. Not only was I bone tired by this time, but I kept running into people I knew. I realized that this was my last opportunity to see them this year, so I never made it past the halfway point in the Expo Hall.
I did notice that the crowds were very thin. Saturday was Family Day, but apparently, not many people wanted to risk venturing out. Even the locals were concerned which is never a good sign.
The Heritage Theater had a full schedule of events, but there were very few people in the audience through no fault of the speakers or RootsTech.
For those hearty souls who did attend, they received up-close and personal sessions and information from the presenters.
It was nice to see the folks from Family Tree Magazine again. I’ve written for them off and on for years, but had never met the staff in person before.
Be sure to check out their Best Genetic Genealogy Websites and also their Genealogy Books Guide, listed by subject.
When you get there, check out their other “best of” categories and other topics.
Walking on down the aisle, I stopped to talk to the “One Kind Act a Day” people,
Being the skeptic that I am, I kept trying to find the hook, but I couldn’t. It’s a nonprofit that seems to do exactly what it says.
This is absolutely something I can sign up for, so I did and took the pledge.
Doing one kind act a day is easy, so let’s do two!
You can follow them on Facebook, too.
Reclaim the Records is another nonprofit that has successfully advocated and reclaimed more than 60 million records to date that were behind lock and key.
Take a look at their successes and their to-do list.
They style themselves as intellectual freedom fighters. Did you know so many records were still entirely unavailable?
Hey, isn’t that Myko Cleland sitting at the Reclaim booth, on the left? He’s the Director of Content in Europe for MyHeritage, nicknamed the DapperHistorian, and you just never know where you’re going to find him!
How cool is this? Wear What You Love uses sublimation dying to permanently print/infuse your photos on t-shirts or other materials.
This also works on fabric that can be used in quilts but more reliably on polyester fabrics, not cotton.
Hmmm, I have some ideas.
Y-DNA Ask Me Anything at FamilyTreeDNA
The Y-DNA “Ask Me Anything” session began at 10:30. I don’t think attendees realized that FamilyTreeDNA brought the R&D brain trust and you could literally ask them anything. What an opportunity!
Left to right, Michael Sager, FamilyTreeDNA’s well-known Y-DNA phylogeneticist, Dr. Paul Maier, seated, population geneticist, and Goran Runfeldt, standing at right, Head of R&D.
The team reviewed how to use Discover and what can be revealed.
Janine Cloud, Manager of Group Projects, is beside me in the black shirt, seated at far right. Group Projects are important tools for Y-DNA testing and testers.
In addition to the Discover Time Tree, shown on the screen above, a Group Time Tree shows Big Y project members as grouped by the volunteer administrators, along with their earliest known ancestors (EKA.)
Here’s an example from the Estes surname project that I administer. My grouping of participants is shown at left, the Time Tree in the center, and the locations with earliest known ancestors at right. Results are displayed in the order that they are phylogenetically related, helping genealogists immensely.
Here, the team is explaining the Block Tree which displays matches in a different format.
Men displayed together on the same Block Tree branch are more closely related to each other than to men displayed in other branches.
Michael Sager observes while Paul Maier demonstrates Globetrekker, an innovative interactive map that shows the path that one’s male ancestors took on their journey from Africa to where they are most recently found.
One of the attendees had a question and looks on as the team explains their results using Globetrekker.
We tried to get a team photo after the presentation and managed to corral some of the team. You’ve met several already, but Bennett Greenspan, Founder and President Emeritus of FamilyTreeDNA, is to my right as you look at the photo, with Sherman McRae standing between Bennett and Paul.
I particularly like this “generations” photo.
In the rear, Katherine Borges stands with Bennett Greenspan. Bennett obviously founded the company, and Katherine was one of the early administrators. Dr. Lior Rauchberger, CEO of myDNA, which includes FamilyTreeDNA, is seated at left, along with Alex Zawisza, CFO, at right. MyDNA purchased Gene by Gene, which includes FamilyTreeDNA, just over three years ago, and the team has continued to work together for the benefit of FamilyTreeDNA customers.
Lior traveled from Australia to attend RootsTech. He could be seen checking people out at the booth, so he had the opportunity to talk with customers. He said he heard the words “brick wall” more in those three days than ever before, as in, “Thanks to FamilyTreeDNA, I broke down my brick wall.”
We all owe Lior a huge debt of gratitude for his continued commitment to FamilyTreeDNA research, and in particular, the Big Y-700 tools, such as Discover, along with the Million Mito Project which will be released with a similar tool, MitoDiscover.
Thanks Lior!
I turned around to see Stephanie Gilbert, who gave the keynote at the FamilyTreeDNA conference.
Stephanie is an incredibly engaging speaker, and I’m going to recommend her to RootsTech for next year.
It was wonderful to see Schelly Talalay Dardashti, at left. She has worked for MyHeritage since 2006 and administers the Tracing the Tribe – Jewish Genealogy Facebook group, which has more than 73,000 members. Schelly is a wonderful ambassador, always helpful and incredibly knowledgeable.
Between us is Dana Stewart Leeds, creator of the Leeds Method, a technique that launched the autocluster craze by manually grouping matches. I wrote about the Leeds Method, here, in 2018. When you see AutoClusters at Genetic Affairs or the Collins-Leeds method at DNAGedcom, think of and thank Dana. They automated her process, with her permission, of course, creating some of the most useful tools available to genealogists. You can follow Dana here.
I swear, it was brainiac day at RootsTech!
Mags Gaulden, one of the founders of mitoYDNA and who writes at Grandma’s Genes, was working in the FamilyTreeDNA booth and was quite busy – so busy that I almost didn’t manage a picture with her. We never did get to have a meal together. We will have to do better in October when we are both scheduled to be at the East Coast Genetic Genealogy Conference in person. Oops, did I say that out loud???
Save the dates!
GEDmatch – New AutoCluster Endogamy Tool
I’ve emailed back and forth with Tom Osypian with GEDmatch many times now, but I’ve never met him in person, even though we’ve been in the same place before.
This time, I was determined. Although Tom was busy several times when I stopped by the booth, there were fewer people on Saturday, so I stood a fighting chance.
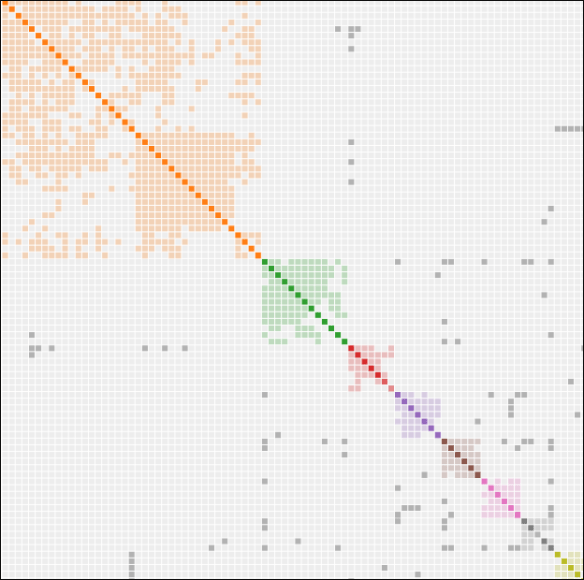
Tom explained that GEDmatch has a new AutoCluster tool developed by Evert-Jan Blom at Genetic Affairs and Jarret Ross from GeneaVlogger that helps with unraveling endogamy. I told him that I already knew because we used my Mom’s autosomal results during testing. Mom is partly endogamous through her grandfather’s Acadian line.
The Acadian cluster in the upper left quadrant looks like an orange blob with no differentiation, where everyone is related to everyone else – because that’s truly how Acadian descendants are connected. As my Acadian cousin once said, “If you’re related to one Acadian, you’re related to all Acadians,” and it’s true.
Evert-Jan needed to optimize clusters for a partially endogamous person without negatively affecting their non-endogamous clusters.
He did a great job separating my Mom’s big orange blob endogamous cluster into these nice, neat mini-clusters.
To take a look, choose AutoCluster Endogamy on GEDmatch and make your preset selection.
There’s a YouTube video about this tool by GeneaVlogger, here.
Next I ran into Patricia Coleman, a fellow genealogist scientist, who wrote an excellent article about finding segment links to the opposite parent using AutoSegment AutoClusters, here. Check out her blog and published papers, here.
We are incredibly fortunate to have such dedicated researchers and scientists in our community.
Unfortunately, I was running out of time on the show floor.
Sisters of Heart
OK, now, I’m going to say something really sappy. Consider yourself warned.
By this time, I needed to find food and quickly eat before my session, which was scheduled to start at 1:15. This meant I needed to be in the room by 12:45.
Janine was doing consultations in the FamilyTreeDNA booth and couldn’t get away for food either.
Thankfully, with the storm approaching, there weren’t long lines at the food vendors. I peeked outside as I walked down the hallway looking for a food booth that wasn’t very busy.
It was ominously dark and gloomy outside, and had begun to snow.
I found the food stand that looked least bad and got in line. Neither Janine nor I knew what was available at the food vendors, but we’ve known each other for enough years and attended enough conferences that we kind of know what the other likes.
I was standing in line taking pictures of the menu and the pre-made foods in the cooler and messaging them to Janine. People must have wondered if I couldn’t find something better to take pictures of. I just chuckled. I’ll spare you the food pictures because they were unremarkable,
They were out of everything Janine thought looked good. Apparently, everyone else thought those items looked good, too. When it was my turn to order, and I had to choose, I messaged Janine that we were sharing a turkey wrap and asked if she wanted fruit.
“YES! Fruit sounds wonderful.”
Great!
I got both items and paid.
“So do chips. Chips sound great, too.”
Perfect.
I paid again.
Then I saw the muffins. Chocolate sour cream swirl muffins with large shiny sugar crystals baked on top.
No need to message Janine about this one.
Yep, I paid for the third time.
Then, I apologized to the people behind me, hoped they didn’t recognize me, and hurried back to the FamilyTreeDNA booth.
Janine’s customer had just finished up, so I sat down in that seat and spread out our goodies on the table between us. The turkey wrap was cut in half, and we shared half of everything.
I love breaking bread and sharing food with my favorite people. There’s something about feeding the body that nourishes the soul and bonds the heart. I can’t explain it, and I really wasn’t thinking about it just then. Both of us just needed a minute to relax and eat before rushing off to do something else.
I asked Janine if she wanted the last part of my half of the turkey wrap. She told me to take the turkey out and eat it because I needed the protein.
Bless her heart. She was right.
I grabbed two forks in the food booth, and we both ate out of the fruit box positioned halfway between us.
Then, after discussing and laughing that the muffin looked like a geode, I cut it into four sections. We ate them on the cupcake paper with forks, like cake. It tasted wonderful. If you’re thinking that I couldn’t finish my turkey wrap, but had plenty of room for chocolate cake, you’d be exactly right!
Someplace in the midst of our impromptu picnic meal, I realized that four years ago at RootsTech 2020, was the last time we would see each other – for years. A week after RootsTech, everything shut down. People died. Both of us had family members who perished in the Covid epidemic.
Everyone was traumatized.
Neither of us knew if we’d ever see each other again, but neither of us verbalized that because – well – we just couldn’t. Some days during that time, it was all any of us could do to simply hold it together.
I realized just how important these very relaxed impromptu moments, built on years of shared space and breaking bread together, really are. It’s exactly why we don’t have any old photos of “normal” things, just special occasions. Normal isn’t special, until it is – when someone is suddenly gone. Then, “normal” is everything.
None of us know which meal together will be the last. We never know when our number will be called, or how. We really only ever have today.
I wish someone had taken a picture of us smiling and eating, sharing our meal with each other, something we’ve done countless times before. Something so normal that we don’t even think about it. I never thought about taking a picture of something so routine, and neither did anyone else. Why would they?
Regardless, that moment is burned into my memory, along with just how precious our time together is.
Then, the moment of quiet respite, eating chocolate muffins and sharing more than food, was over, and the fragile thought bubble was broken by the ticking of the clock. I had to jump up and run off to my next presentation, and a customer approached and asked Janine a question.
Thank Goodness we were both able to return to RootsTech and relish something so absolutely normal once again.
Highways of History – Flesh Out Your Ancestors Using Discover Case Studies
My class on Friday, “DNA Academy,” was full, and sadly, people were being turned away at the door. Saturday’s “Highways of History” class was held in a larger room, but many people stayed home, so the room was only about three-quarters full. I forgot to ask someone to take a picture, so I’ll just share a few slides.
I really enjoy using AI occasionally for images. This was ChatGPTs idea of Highways of History.
Using Big Y DNA results, I provided examples of using the Discover tools to reveal the stories of my ancestors. Not every Discover tool reveals something amazing about each ancestor, but together, they tell a story we can’t unravel any other way.
I seek out men who descend from every male ancestor paternally through all males and offer a scholarship for Big Y-700 testing.
Here are just a few examples of what I’ve found and documented:
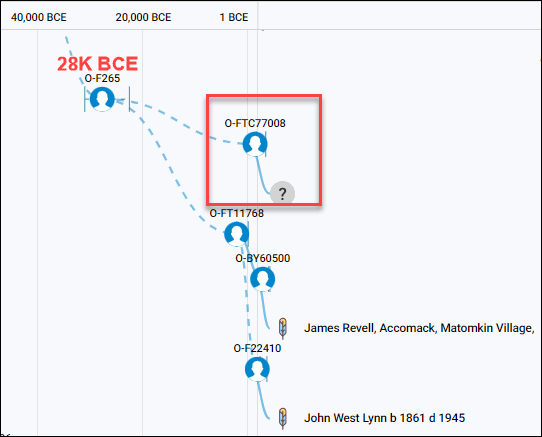
- A descendant of Etienne Hebert (c1626-c1670), my Acadian ancestor, matches an ancient DNA burial found in Metz, France. Etienne and his brother’s children cluster in a group with a common ancestor about 1650, and the ancient burial dates to about the year 500 CE during the time that Metz was a Gallo Celtic Village. Among other things, we learn that their common ancestors were Celtic.
- An adopted male matches several Estes men. Based on his Big Y-700 mutations, I can place him in the Estes family tree within two generations. His position in the tree is confirmed by autosomal matches to the ancestors of the wife of Joseph Frank Estes. Autosomal matches confirmed the Big Y-700.
- Germain Doucet, born in France in the late 1500s, had two sons. One was born in France about 1621, and the second in Acadia (now Nova Scotia) was born to either a second or third wife in 1641 and named after Germain. Based on Big Y-700 tests, the son born in 1621 has a European haplogroup, but Germain, born in 1641, has a Native American father, suggesting the possibility that he may have been adopted by the older Germain Doucet. This was quite an unexpected surprise.
- A Bowling descendant of Hugh Bowling (1591-1651) born in Chorley, Lancashire, England, had almost no English matches. STR matches are from Saudi Arabia, Algeria, Cyprus, Germany, and Portugal, but the highest percentage are from Spain. Furthermore, his ancient Connections are from Hungary, Israel (4), Armenia, Rome, Italy, Turkey, Lebanon, Lincolnshire, and Norwich, England. Local history reveals a Roman Fort just 19 miles away from where Hugh Bowling lived, and the location, now excavated, was a settlement location for Roman Sarmatian soldiers.
- Thomas Speak was born about 1634 in Downham, Lancashire, near Chorley, England where the family attended church. Big Y-700 testing shows that he and other English Speak men still living in the area share an ancestor about 1300 CE. When we visited in 2012, we discovered that Myles Standish’s family also attended the same church. Saxon Crosses are found in the graveyard outside, dating to circa 800-900 CE. A Standish male’s Big Y-700 test matches the Speak men, with their common ancestor dating to 850 CE, the same time that the Saxons were settling the region.
- Bennett Greenspan’s Jewish ancestors were found in Ukraine in the mid-1700s, but he wanted to know more about where they came from originally. Were they Ashkenazi or Sephardic, or something else? By upgrading both close and distant matches to the Big Y-700, Bennett discovered that their common ancestors were in Spain in the year 296 when the two lines diverged and his line left. You can read Bennett’s story in more detail, here.
None of these mysteries or brick walls could have been solved without Big Y-700 tests and without the Discover tools.
This session was so much fun, and I can hardly wait to find more male ancestors and test their direct male-line descendants.
Goodbyes
By the time questions were answered, and I packed up my equipment, there were only about 15 minutes left until the Expo Hall closed at 3. Furthermore, I needed to retrieve my coat from the FamilyTreeDNA booth, retrieve my suitcase from the Marriott bellman, and order an Uber. My flight was only about two and a half hours away, assuming it left.
So far, it hadn’t been cancelled or delayed.
I mentioned my flight concerns to a colleague that I ran into on the way to the booth. He happens to live in Salt Lake City and gave me his phone number, with instructions to call if I got stranded.
My first (unspoken) thought was, “Thank you, but I’d never impose like that.” But then, I realized that was crazy and I really should call him if I needed help. What was wrong with me? I didn’t know him well, but I had known him and the company where he works for many years and felt completely safe. We are Facebook friends too, so I’ve joyfully watched him marry and start a family. I would have done exactly the same for him, and yes, I absolutely WOULD have wanted him to call. Plus, if I actually did wind up staying on his couch for a day or so, I would get time to “Grandma” his children, so HUGE BONUS!
You know who you are, and THANK YOU. I felt so much better after that. Genealogists are just the most amazing people!
Then, I ran into Lisa Rhea Baker who very generously gifted me with bracelets made by her veteran daughter as she healed from surgery. The bracelets around my wrist are beaded, and the one joining our hands is knotted in German colors. I’m wearing that one today. What a very talented and generous young lady.
I was very touched and so grateful. I asked her to thank her daughter on my behalf.
I saw Katherine Borges again in the booth as I was retrieving my coat and we quickly took a selfie. Neither of us realized we hadn’t gotten one earlier, although we did manage to have dinner with a small group where we all chattered like magpies.
Last, Goran, Paul and I took a quick selfie as I was preparing to run out the door. It was 3, closing time, and almost no one was left in the Expo Hall. I knew if I missed this flight, I’d not get another one. Everything was full.
I surely miss seeing these guys. Hopefully, I’ll see them again before the next RootsTech!
The Blizzard Strikes
I stepped outside.
The blizzard had begun in earnest. I could see a couple blocks down the street, but huge flakes of snow were pouring down. The wind was blowing viciously, whipping everything, making it difficult to hang onto my laptop rolling bag. The snow was sticking to everything.
At least it wasn’t slick yet, at least not where I was walking. If the wind hadn’t been so strong, it would have been pretty.
Would the plane be able to take off in this wind? The snow was blowing directly sideways now.
The only distance I had to walk was across the street. This is how much snow accumulated on my coat in just a minute or so.
A little later, Goran took this picture.
Ubers were becoming somewhat scarce, so two of us shared and made it to the airport in time for long TSA lines.
The plane was about 45 minutes late, which didn’t surprise me. I heaved a huge sigh of relief when it pulled up to the gate. At least it arrived, and as soon as it was cleaned a bit, we began to board.
Eventually, we pulled out of the gate and began waiting on the tarmac for the plane to be de-iced.
An hour later, we weren’t even halfway to the front of the line. The pilot estimated it would be another 90 minutes or so.
The snow continued to accumulate.
Would the pilot and crew time out and be unable to fly?
If we had to go back, there would be no prayer of getting another crew. Flights were already being canceled.
The woman beside me was ill. I felt awful for her, and it occurred to me that this might also be a reason to return to the gate.
At least the pilot allowed us to unbuckle our seatbelts and go to the restroom as we waited.
My flight had been scheduled to arrive just after midnight. But now, we were more than four hours late. What time would we get in? My poor husband. I told him to go to sleep and I’d just stay in the hotel in the airport. He said no, nothing doing.
I begged him to at least take a nap and recheck the flights at 3 or 4 AM.
The flight was extremely rough. We couldn’t get above or around the storm, and the seatbelt sign was only off for about 10 minutes during the entire flight.
I tried to sleep, but that wasn’t happening, even though I was beyond exhausted.
This is what love looks like. One single car in the cell lot at around 5 AM, as Jim waited patiently for me.
On the way home, in fact, all of the way home, we drove through the most incredible lightning storm I’ve ever seen.
It was someplace between worrisome/terrifying, and fascinating.
This lightning wasn’t reaching toward the earth in bolts. Instead, the entire sky lit up like daylight, horizon to horizon, flashing like an extremely bright strobe. It was so bright that, at times, it was nearly blinding, and the clouds looked like rainbows as the lightning flashed behind and through them. I had never seen anything like this.
This type of “sheet lightning” is crazy rare. Thankfully, it kept us awake and was stunningly beautiful in a very strange, ethereal way. We worried that we would be caught in a hellacious storm and unable to see in the downpour.
Florida is notorious for vicious storms and torrential downpours. It’s also the lightning strike capital of the US and ranks fourth in the world. This area, in particular, is known as Lightning Alley. Our house was struck last year.
As we exited the expressway, just a couple miles from home, the sky unzipped, and torrential rains began. Thankfully, we were spared for most of the drive.
I was incredibly glad to finally be home and hoped that others had been able to either escape the Utah storm or find a room in a hotel that did not lose power on Sunday. Reports said wind gusts in the Utah mountains were measured at 165 miles an hour, but Salt Lake City, tucked into a valley, was spared most of that.
What an incredible week in so very many ways.
I hope you enjoyed coming along with me. Dates have already been announced for RootsTech 2025.
_____________________________________________________________
Follow DNAexplain on Facebook, here.
Share the Love!
You’re always welcome to forward articles or links to friends and share on social media.
If you haven’t already subscribed (it’s free,) you can receive an e-mail whenever I publish by clicking the “follow” button on the main blog page, here.
You Can Help Keep This Blog Free
I receive a small contribution when you click on some of the links to vendors in my articles. This does NOT increase your price but helps me keep the lights on and this informational blog free for everyone. Please click on the links in the articles or to the vendors below if you are purchasing products or DNA testing.
Thank you so much.
DNA Purchases and Free Uploads
- FamilyTreeDNA – Y, mitochondrial and autosomal DNA testing
- MyHeritage DNA – Autosomal DNA test
- MyHeritage FREE DNA file upload – Upload your DNA file from other vendors free
- AncestryDNA – Autosomal DNA test
- AncestryDNA Plus Traits
- 23andMe Ancestry – Autosomal DNA only, no Health
- 23andMe Ancestry Plus Health
Genealogy Products and Services
- MyHeritage Subscription with Free Trial
- Legacy Family Tree Webinars – Genealogy and DNA classes, subscription-based, some free
- Legacy Family Tree Software – Genealogy software for your computer
- OldNews – Old Newspapers with links to save to MyHeritage trees
- Newspapers.com – Search newspapers for your ancestors
- NewspaperArchive – Search different newspapers for your ancestors
My Book
- DNA for Native American Genealogy – by Roberta Estes, for those ordering the e-book from anyplace, or paperback within the United States
- DNA for Native American Genealogy – for those ordering the paperback outside the US
Genealogy Books
- Genealogical.com – Lots of wonderful genealogy research books
- American Ancestors – Wonderful selection of genealogy books
Genealogy Research
- Legacy Tree Genealogists – Professional genealogy research