Many males take the Big Y-700 test offered by FamilyTreeDNA, so named because testers receive the most granular haplogroup SNP results in addition to 700+ included STR marker results. If you’re not familiar with those terms, you might enjoy the article, STRs vs SNPs, Multiple DNA Personalities.
The Big Y test gives testers the best of both, along with contributing to the building of the Y phylotree. You can read about the additions to the Y tree via the Big Y, plus how it helped my own Estes project, here.
Some men order this test of their own volition, some at the request of a family member, and some in response to project administrators who are studying a specific topic – like a particular surname.
The Big Y-700 test is the most complete Y DNA test offered, testing millions of locations on the Y chromosome to reveal mutations, some unique and never before discovered, many of which are useful to genealogists. The Big Y-700 includes the traditional Y DNA STR marker testing along with SNP results that define haplogroups. Translated, both types of test results are compared to other men for genealogy, which is the primary goal of DNA testing.
Being a female, I often recruit males in my family surname lines and sponsor testing. My McNiel line, historic haplogroup R-M222, has been particularly frustrating both genealogically as well as genetically after hitting a brick wall in the 1700s. My McNeill cousin agreed to take a Big Y test, and this analysis walks through the process of understanding what those results are revealing.
After my McNeill cousin’s Big Y results came back from the lab, I spent a significant amount of time turning over every leaf to extract as much information as possible, both from the Big Y-700 DNA test itself and as part of a broader set of intertwined genetic information and genealogical evidence.
I invite you along on this journey as I explain the questions we hoped to answer and then evaluate Big Y DNA results along with other information to shed light on those quandaries.
I will warn you, this article is long because it’s a step-by-step instruction manual for you to follow when interpreting your own Big Y results. I’d suggest you simply read this article the first time to get a feel for the landscape, before working through the process with your own results. There’s so much available that most people leave laying on the table because they don’t understand how to extract the full potential of these test results.
If you’d like to read more about the Big Y-700 test, the FamilyTreeDNA white paper is here, and I wrote about the Big Y-700 when it was introduced, here.
You can read an overview of Y DNA, here, and Y DNA: The Dictionary of DNA, here.
Ok, get yourself a cuppa joe, settle in, and let’s go!
George and Thomas McNiel – Who Were They?
George and Thomas McNiel appear together in Spotsylvania County, Virginia records. Y DNA results, in combination with early records, suggest that these two men were brothers.
I wrote about discovering that Thomas McNeil’s descendant had taken a Y DNA test and matched George’s descendants, here, and about my ancestor George McNiel, here.
McNiel family history in Wilkes County, NC, recorded in a letter written in 1898 by George McNiel’s grandson tells us that George McNiel, born about 1720, came from Scotland with his two brothers, John and Thomas. Elsewhere, it was reported that the McNiel brothers sailed from Glasgow, Scotland and that George had been educated at the University of Edinburgh for the Presbyterian ministry but had a change of religious conviction during the voyage. As a result, a theological tiff developed that split the brothers.
George, eventually, if not immediately, became a Baptist preacher. His origins remain uncertain.
The brothers reportedly arrived about 1750 in Maryland, although I have no confirmation. By 1754, Thomas McNeil appeared in the Spotsylvania County, VA records with a male being apprenticed to him as a tailor. In 1757, in Spotsylvania County, the first record of George McNeil showed James Pey being apprenticed to learn the occupation of tailor.
If George and Thomas were indeed tailors, that’s not generally a country occupation and would imply that they both apprenticed as such when they were growing up, wherever that was.
Thomas McNeil is recorded in one Spotsylvania deed as being from King and Queen County, VA. If this is the case, and George and Thomas McNiel lived in King and Queen, at least for a time, this would explain the lack of early records, as King and Queen is a thrice-burned county. If there was a third brother, John, I find no record of him.
My now-deceased cousin, George McNiel, initially tested for the McNiel Y DNA and also functioned for decades as the family historian. George, along with his wife, inventoried the many cemeteries of Wilkes County, NC.
George believed through oral history that the family descended from the McNiel’s of Barra.

George had this lovely framed print of Kisimul Castle, seat of the McNiel Clan on the Isle of Barra, proudly displayed on his wall.
That myth was dispelled with the initial DNA testing when our line did not match the Barra line, as can be seen in the MacNeil DNA project, much to George’s disappointment. As George himself said, the McNiel history is both mysterious and contradictory. Amen to that, George!

However, in place of that history, we were instead awarded the Niall of the 9 Hostages badge, created many years ago based on a 12 marker STR result profile. Additionally, the McNiel DNA was assigned to haplogroup R-M222. Of course, today’s that’s a far upstream haplogroup, but 15+ years ago, we had only a fraction of the testing or knowledge that we do today.
The name McNeil, McNiel, or however you spell it, resembles Niall, so on the surface, this made at least some sense. George was encouraged by the new information, even though he still grieved the loss of Kisimul Castle.
Of course, this also caused us to wonder about the story stating our line had originated in Scotland because Niall of the 9 Hostages lived in Ireland.
Niall of the 9 Hostages
Niall of the 9 Hostages was reportedly a High King of Ireland sometime between the 6th and 10th centuries. However, actual historical records place him living someplace in the mid-late 300s to early 400s, with his death reported in different sources as occurring before 382 and alternatively about 411. The Annals of the Four Masters dates his reign to 379-405, and Foras Feasa ar Eirinn says from 368-395. Activities of his sons are reported between 379 and 405.
In other words, Niall lived in Ireland about 1500-1600 years ago, give or take.
Migration
Generally, migration was primarily from Scotland to Ireland, not the reverse, at least as far as we know in recorded history. Many Scottish families settled in the Ulster Plantation beginning in 1606 in what is now Northern Ireland. The Scots-Irish immigration to the states had begun by 1718. Many Protestant Scottish families immigrated from Ireland carrying the traditional “Mc” names and Presbyterian religion, clearly indicating their Scottish heritage. The Irish were traditionally Catholic. George could have been one of these immigrants.
We have unresolved conflicts between the following pieces of McNeil history:
- Descended from McNeil’s of Barra – disproved through original Y DNA testing.
- Immigrated from Glasgow, Scotland, and schooled in the Presbyterian religion in Edinburgh.
- Descended from the Ui Neill dynasty, an Irish royal family dominating the northern half of Ireland from the 6th to 10th centuries.
Of course, it’s possible that our McNiel/McNeil line could have been descended from the Ui Neill dynasty AND also lived in Scotland before immigrating.
It’s also possible that they immigrated from Ireland, not Scotland.
And finally, it’s possible that the McNeil surname and M222 descent are not related and those two things are independent and happenstance.
A New Y DNA Tester
Since cousin George is, sadly, deceased, we needed a new male Y DNA tester to represent our McNiel line. Fortunately, one such cousin graciously agreed to take the Big Y-700 test so that we might, hopefully, answer numerous questions:
- Does the McNiel line have a unique haplogroup, and if so, what does it tell us?
- Does our McNiel line descend from Ireland or Scotland?
- Where are our closest geographic clusters?
- What can we tell by tracing our haplogroup back in time?
- Do any other men match the McNiel haplogroup, and what do we know about their history?
- Does the Y DNA align with any specific clans, clan history, or prehistory contributing to clans?
With DNA, you don’t know what you don’t know until you test.
Welcome – New Haplogroup
I was excited to see my McNeill cousin’s results arrive. He had graciously allowed me access, so I eagerly took a look.
He had been assigned to haplogroup R-BY18350.

Initially, I saw that indeed, six men matched my McNeill cousin, assigned to the same haplogroup. Those surnames were:
- Scott
- McCollum
- Glass
- McMichael
- Murphy
- Campbell
Notice that I said, “were.” That’s right, because shortly after the results were returned, based on markers called private variants, Family Tree DNA assigned a new haplogroup to my McNeill cousin.
Drum roll please!!!
Haplogroup R-BY18332

Additionally, my cousin’s Big Y test resulted in several branches being split, shown on the Block Tree below.

How cool is this!
This Block Tree graphic shows, visually, that our McNiel line is closest to McCollum and Campbell testers, and is a brother clade to those branches showing to the left and right of our new R-BY18332. It’s worth noting that BY25938 is an equivalent SNP to BY18332, at least today. In the future, perhaps another tester will test, allowing those two branches to be further subdivided.
Furthermore, after the new branches were added, Cousin McNeill has no more Private Variants, which are unnamed SNPs. There were all utilized in naming additional tree branches!
I wrote about the Big Y Block Tree here.
Niall (Or Whoever) Was Prolific
The first thing that became immediately obvious was how successful our progenitor was.

click to enlarge
In the MacNeil DNA project, 38 men with various surname spellings descend from M222. There are more in the database who haven’t joined the MacNeil project.
Whoever originally carried SNP R-M222, someplace between 2400 and 5900 years ago, according to the block tree, either had many sons who had sons, or his descendants did. One thing is for sure, his line certainly is in no jeopardy of dying out today.
The Haplogroup R-M222 DNA Project, which studies this particular haplogroup, reads like a who’s who of Irish surnames.
Big Y Match Results
Big Y matches must have no more than 30 SNP differences total, including private variants and named SNPs combined. Named SNPs function as haplogroup names. In other words, Cousin McNeill’s terminal SNP, meaning the SNP furthest down on the tree, R-BY18332, is also his haplogroup name.
Private variants are mutations that have occurred in the line being tested, but not yet in other lines. Occurrences of private variants in multiple testers allow the Private Variant to be named and placed on the haplotree.
Of course, Family Tree DNA offers two types of Y DNA testing, STR testing which is the traditional 12, 25, 37, 67 and 111 marker testing panels, and the Big Y-700 test which provides testers with:
- All 111 STR markers used for matching and comparison
- Another 589+ STR markers only available through the Big Y test increasing the total STR markers tested from 111 to minimally 700
- A scan of the Y chromosome, looking for new and known SNPs and STR mutations
Of course, these tests keep on giving, both with matching and in the case of the Big Y – continued haplogroup discovery and refinement in the future as more testers test. The Big Y is an investment as a test that keeps on giving, not just a one-time purchase.
I wrote about the Big Y-700 when it was introduced here and a bit later here.
Let’s see what the results tell us. We’ll start by taking a look at the matches, the first place that most testers begin.

Regular Y DNA STR matching shows the results for the STR results through 111 markers. The Big Y section, below, provides results for the Big Y SNPs, Big Y matches and additional STR results above 111 markers.

Let’s take a look.
STR and SNP Testing
Of Cousin McNeil’s matches, 2 Big Y testers and several STR testers carry some variant of the Neal, Neel, McNiel, McNeil, O’Neil, etc. surnames by many spellings.
While STR matching is focused primarily on a genealogical timeframe, meaning current to roughly 500-800 years in the past, SNP testing reaches much further back in time.
- STR matching reaches approximately 500-800 years.
- Big Y matching reaches approximately 1500 years.
- SNPs and haplogroups reach back infinitely, and can be tracked historically beyond the genealogical timeframe, shedding light on our ancestors’ migration paths, helping to answer the age-old question of “where did we come from.”
These STR and Big Y time estimates are based on a maximum number of mutations for testers to be considered matches paired with known genealogy.
Big Y results consider two men a match if they have 30 or fewer total SNP differences. Using NGS (next generation sequencing) scan technology, the targeted regions of the Y chromosome are scanned multiple times, although not all regions are equally useful.
Individually tested SNPs are still occasionally available in some cases, but individual SNP testing has generally been eclipsed by the greatly more efficient enriched technology utilized with Big Y testing.
Think of SNP testing as walking up to a specific location and taking a look, while NGS scan technology is a drone flying over the entire region 30-50 times looking multiple times to be sure they see the more distant target accurately.
Multiple scans acquiring the same read in the same location, shown below in the Big Y browser tool by the pink mutations at the red arrow, confirm that NGS sequencing is quite reliable.

These two types of tests, STR panels 12-111 and the SNP-based Big Y, are meant to be utilized in combination with each other.
STR markers tend to mutate faster and are less reliable, experiencing frustrating back mutations. SNPs very rarely experience this level of instability. Some regions of the Y chromosome are messier or more complicated than others, causing problems with interpreting reads reliably.
For purposes of clarity, the string of pink A reads above is “not messy,” and “A” is very clearly a mutation because all ~39 scanned reads report the same value of “A,” and according to the legend, all of those scans are high quality. Multiple combined reads of A and G, for example, in the same location, would be tough to call accurately and would be considered unreliable.
You can see examples of a few scattered pink misreads, above.
The two different kinds of tests produce results for overlapping timeframes – with STR mutations generally sifting through closer relationships and SNPs reaching back further in time.
Many more men have taken the Y DNA STR tests over the last 20 years. The Big Y tests have only been available for the past handful of years.
STR testing produces the following matches for my McNiel cousin:
| STR Level |
STR Matches |
STR Matches Who Took the Big Y |
% STR Who Took Big Y |
STR Matches Who Also Match on the Big Y |
| 12 |
5988 |
796 |
13 |
52 |
| 25 |
6660 |
725 |
11 |
57 |
| 37 |
878 |
94 |
11 |
12 |
| 67 |
1225 |
252 |
21 |
23 |
| 111 |
4 |
2 |
50 |
1 |
Typically, one would expect that all STR matches that took the Big Y would match on the Big Y, since STR results suggest relationships closer in time, but that’s not the case.
- Many STR testers who have taken the Big Y seem to be just slightly too distant to be considered a Big Y match using SNPs, which flies in the face of conventional wisdom.
- However, this could easily be a function of the fact that STRs mutate both backward and forwards and may have simply “happened” to have mutated to a common value – which suggests a closer relationship than actually exists.
- It could also be that the SNP matching threshold needs to be raised since the enhanced and enriched Big Y-700 technology now finds more mutations than the older Big Y-500. I would like to see SNP matching expanded to 40 from 30 because it seems that clan connections may be being missed. Thirty may have been a great threshold before the more sensitive Big Y-700 test revealed more mutations, which means that people hit that 30 threshold before they did with previous tests.
- Between the combination of STRs and SNPs mutating at the same time, some Big Y matches are pushed just out of range.
In a nutshell, the correlation I expected to find in terms of matching between STR and Big Y testing is not what I found. Let’s take a look at what we discovered.
It’s worth noting that the analysis is easier if you are working together with at least your closest matches or have access via projects to at least some of their results. You can see common STR values to 111 in projects, such as surname projects. Project administrators can view more if project members have allowed access.
Unexpected Discoveries and Gotchas
While I did expect STR matches to also match on the Big Y, I don’t expect the Big Y matches to necessarily match on the STR tests. After all, the Big Y is testing for more deep-rooted history.
Only one of the McNiel Big Y matches also matches at all levels of STR testing. That’s not surprising since Big Y matching reaches further back in time than STR testing, and indeed, not all STR testers have taken a Big Y test.
Of my McNeill cousin’s closest Big Y matches, we find the following relative to STR matching.
| Surname |
Ancestral Location |
Big Y Variant/SNP Difference |
STR Match Level |
| Scott |
1565 in Buccleuch, Selkirkshire, Scotland |
20 |
12, 25, 37, 67 |
| McCollum |
Not listed |
21 |
67 only |
| Glass |
1618 in Banbridge, County Down, Ireland |
23 |
12, 25, 67 |
| McMichael |
1720 County Antrim, Ireland |
28 |
67 only |
| Murphy |
Not listed |
29 |
12, 25, 37, 67 |
| Campbell |
Scotland |
30 |
12, 25, 37, 67, 111 |
It’s ironic that the man who matches on all STR levels has the most variants, 30 – so many that with 1 more, he would not have been considered a Big Y match at all.
Only the Campbell man matches on all STR panels. Unfortunately, this Campbell male does not match the Clan Campbell line, so that momentary clan connection theory is immediately put to rest.
Block Tree Matches – What They Do, and Don’t, Mean
Note that a Carnes male, the other person who matches my McNeill cousin at 111 STR markers and has taken a Big Y test does not match at the Big Y level. His haplogroup BY69003 is located several branches up the tree, with our common ancestor, R-S588, having lived about 2000 years ago. Interestingly, we do match other R-S588 men.
This is an example where the total number of SNP mutations is greater than 30 for these 2 men (McNeill and Carnes), but not for my McNeill cousin compared with other men on the same S588 branch.

By searching for Carnes on the block tree, I can view my cousin’s match to Mr. Carnes, even though they don’t match on the Big Y. STR matches who have taken the Big Y test, even if they don’t match at the Big Y level, are shown on the Block Tree on their branch.
By clicking on the haplogroup name, R-BY69003, above, I can then see three categories of information about the matches at that haplogroup level, below.

click to enlarge
By selecting “Matches,” I can see results under the column, “Big Y.” This does NOT mean that the tester matches either Mr. Carnes or Mr. Riker on the Big Y, but is telling me that there are 14 differences out of 615 STR markers above 111 markers for Mr. Carnes, and 8 of 389 for Mr. Riker.
In other words, this Big Y column is providing STR information, not indicating a Big Y match. You can’t tell one way or another if someone shown on the Block Tree is shown there because they are a Big Y match or because they are an STR match that shares the same haplogroup.
As a cautionary note, your STR matches that have taken the Big Y ARE shown on the block tree, which is a good thing. Just don’t assume that means they are Big Y matches.
The 30 SNP threshold precludes some matches.
My research indicates that the people who match on STRs and carry the same haplogroup, but don’t match at the Big Y level, are every bit as relevant as those who do match on the Big Y.

If you’re not vigilant when viewing the block tree, you’ll make the assumption that you match all of the people showing on the Block Tree on the Big Y test since Block Tree appears under the Big Y tools. You have to check Big Y matches specifically to see if you match people shown on the Block Tree. You don’t necessarily match all of them on the Big Y test, and vice versa, of course.
You match Block Tree inhabitants either:
- On the Big Y, but not the STR panels
- On the Big Y AND at least one level of STRs between 12 and 111, inclusive
- On STRs to someone who has taken the Big Y test, but whom you do not match on the Big Y test
Big Y-500 or Big Y-700?

click to enlarge
Looking at the number of STR markers on the matches page of the Block Tree for BY69003, above, or on the STR Matches page is the only way to determine whether or not your match took the Big Y-700 or the Big Y-500 test.
If you add 111 to the Big Y SNP number of 615 for Mr. Carnes, the total equals 726, which is more than 700, so you know he took the Big Y-700.
If you add 111 to 389 for Mr. Riker, you get 500, which is less than 700, so you know that he took the Big Y-500 and not the Big Y-700.
There are still a very small number of men in the database who did not upgrade to 111 when they ordered their original Big Y test, but generally, this calculation methodology will work. Today, all Big Y tests are upgraded to 111 markers if they have not already tested at that level.
Why does Big Y-500 vs Big Y-700 matter? The enriched chemistry behind the testing technology improved significantly with the Big Y-700 test, enhancing Y-DNA results. I was an avowed skeptic until I saw the results myself after upgrading men in the Estes DNA project. In other words, if Big Y-500 testers upgrade, they will probably have more SNPs in common.
You may want to contact your closest Big Y-500 matches and ask if they will consider upgrading to the Big Y-700 test. For example, if we had close McNiel or similar surname matches, I would do exactly that.
Matching Both the Big Y and STRs – No Single Source
There is no single place or option to view whether or not you match someone BOTH on the Big Y AND STR markers. You can see both match categories individually, of course, but not together.
You can determine if your STR matches took the Big Y, below, and their haplogroup, which is quite useful, but you can’t tell if you match them at the Big Y level on this page.

click to enlarge
Selecting “Display Only Matches With Big Y” means displaying matches to men who took the Big Y test, not necessarily men you match on the Big Y. Mr. Conley, in the example above, does not match my McNeill cousin on the Big Y but does match him at 12 and 25 STR markers.
I hope FTDNA will add three display options:
- Select only men that match on the Big Y in the STR panel
- Add an option for Big Y on the advanced matches page
- Indicate men who also match on STRs on the Big Y match page
It was cumbersome and frustrating to have to view all of the matches multiple times to compile various pieces of information in a separate spreadsheet.
No Big Y Match Download
There is also no option to download your Big Y matches. With a few matches, this doesn’t matter, but with 119 matches, or more, it does. As more people test, everyone will have more matches. That’s what we all want!
What you can do, however, is to download your STR matches from your match page at levels 12-111 individually, then combine them into one spreadsheet. (It would be nice to be able to download them all at once.)

You can then add your Big Y matches manually to the STR spreadsheet, or you can simply create a separate Big Y spreadsheet. That’s what I chose to do after downloading my cousin’s 14,737 rows of STR matches. I told you that R-M222 was prolific! I wasn’t kidding.
This high number of STR matches also perfectly illustrates why the Big Y SNP results were so critical in establishing the backbone relationship structure. Using the two tools together is indispensable.
An additional benefit to downloading STR results is that you can sort the STR spreadsheet columns in surname order. This facilitates easily spotting all spelling variations of McNiel, including words like Niel, Neal and such that might be relevant but that you might not notice otherwise.
Creating a Big Y Spreadsheet
My McNiel cousin has 119 Big Y-700 matches.
I built a spreadsheet with the following columns facilitating sorting in a number of ways, with definitions as follows:

click to enlarge
- First Name
- Last Name – You will want to search matches on your personal page at Family Tree DNA by this surname later, so be sure if there is a hyphenated name to enter it completely.
- Haplogroup – You’ll want to sort by this field.
- Convergent – A field you’ll complete when doing your analysis. Convergence is the common haplogroup in the tree shared by you and your match. In the case of the green matches above, which are color-coded on my spreadsheet to indicate the closest matches with my McNiel cousin, the convergent haplogroup is BY18350.
- Common Tree Gen – This column is the generations on the Block Tree shown to this common haplogroup. In the example above, it’s between 9 and 14 SNP generations. I’ll show you where to gather this information.
- Geographic Location – Can be garnered from 4 sources. No color in that cell indicates that this information came from the Earliest Known Ancestor (EKA) field in the STR matches. Blue indicates that I opened the tree and pulled the location information from that source. Orange means that someone else by the same surname whom the tester also Y DNA matches shows this location. I am very cautious when assigning orange, and it’s risky because it may not be accurate. A fourth source is to use Ancestry, MyHeritage, or another genealogical resource to identify a location if an individual provides genealogical information but no location in the EKA field. Utilizing genealogy databases is only possible if enough information is provided to make a unique identification. John Smith 1700-1750 won’t do it, but Seamus McDougal (1750-1810) married to Nelly Anderson might just work.
- STR Match – Tells me if the Big Y match also matches on STR markers, and if so, which ones. Only the first 111 markers are used for matching. No STR match generally means the match is further back in time, but there are no hard and fast rules.
- Big Y Match – My original goal was to combine this information with the STR match spreadsheet. If you don’t wish to combine the two, then you don’t need this column.
- Tree – An easy way for me to keep track of which matches do and do not have a tree. Please upload or create a tree.
You can also add a spreadsheet column for comments or contact information.

You will also want to click your match’s name to display their profile card, paying particular attention to the “About Me” information where people sometimes enter genealogical information. Also, scan the Ancestral Surnames where the match may enter a location for a specific surname.
Private Variants
I added additional spreadsheet columns, not shown above, for Private Variant analysis. That level of analysis is beyond what most people are interested in doing, so I’m only briefly discussing this aspect. You may want to read along, so you at least understand what you are looking at.
Clicking on Private Variants in your Big Y Results shows your variants, or mutations, that are unnamed as SNPs. When they are named, they become SNPs and are placed on the haplotree.
The reference or “normal” state for the DNA allele at that location is shown as the “Reference,” and “Genotype” is the result of the tester. Reference results are not shown for each tester, because the majority are the same. Only mutations are shown.

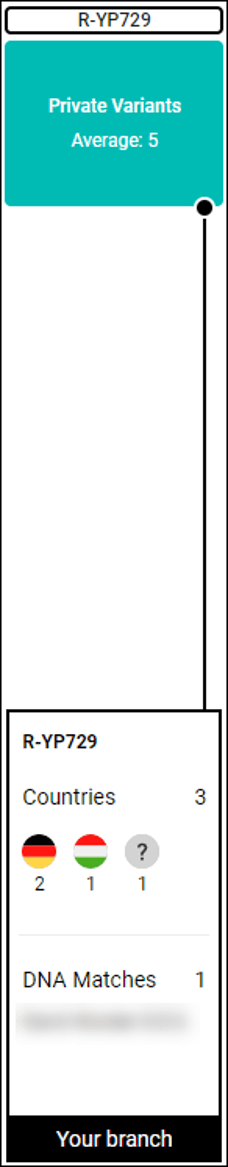
There are 5 Private Variants, total, for my cousin. I’ve obscured the actual variant numbers and instead typed in 111111 and 222222 for the first two as examples.

In our example, there are 6 Big Y matches, with matches one and five having the non-matching variants shown above.
Non-matching variants mean that the match, Mr. Scott, in example 1, does NOT match the tester (my cousin) on those variants.
- If the tester (you) has no mutation, you won’t have a Private Variant shown on your Private Variant page.
- If the tester does have a Private Variant shown, and that variant shows ON their matches list of non-matching variants, it means the match does NOT match the tester, and either has the normal reference value or a different mutation. Explained another way, if you have a mutation, and that variant is listed on your match list of Non-Matching Variants, your match does NOT match you and does NOT have the same mutation.
- If the match does NOT have the Private Variant on their list, that means the match DOES match the tester, and they both have the same mutation, making this Private Variant a candidate to be named as a new SNP.
- If you don’t have a Private Variant listed, but it shows in the Non-Matching Variants of your match, that means you have the reference or normal value, and they have a mutation.
In example #1, above, the tester has a mutation at variant 111111, and 111111 is shown as a Non-Matching Variant to Mr. Scott, so Mr. Scott does NOT match the tester. Mr. Scott also does NOT match the tester at locations 222222 and 444444.
In example #5, 111111 is NOT shown on the Non-Matching Variant list, so Mr. Treacy DOES match the tester.
I have a terrible time wrapping my head around the double negatives, so it’s critical that I make charts.
On the chart below, I’ve listed the tester’s private variants in an individual column each, so 111111, 222222, etc.
For each match, I’ve copy and pasted their Non-Matching Variants in a column to the right of the tester’s variants, in the lavender region. In this example, I’ve typed the example variants into separate columns for each tester so you can see the difference. Remember, a non-matching variant means they do NOT match the tester’s mutation.

On my normal spreadsheet where the non-matching variants don’t have individuals columns, I then search for the first variant, 111111. If the variant does appear in the list, it means that match #1 does NOT have the mutation, so I DON’T put an X in the box for match #1 under 111111.
In the example above, the only match that does NOT have 111111 on their list of Non-Matching Variants is #5, so an X IS placed in that corresponding cell. I’ve highlighted that column in yellow to indicate this is a candidate for a new SNP.
You can see that no one else has the variant, 222222, so it truly is totally private. It’s not highlighted in yellow because it’s not a candidate to be a new SNP.
Everyone shares mutation 333333, so it’s a great candidate to become a new SNP, as is 555555.
Match #6 shares the mutation at 444444, but no one else does.
This is a manual illustration of an automated process that occurs at Family Tree DNA. After Big Y matches are returned, automated software creates private variant lists of potential new haplogroups that are then reviewed internally where SNPs are evaluated, named, and placed on the tree if appropriate.
If you follow this process and discover matches, you probably don’t need to do anything, as the automated review process will likely catch up within a few days to weeks.
Big Y Matches
In the case of the McNiel line, it was exciting to discover several private variants, mutations that were not yet named SNPs, found in several matches that were candidates to be named as SNPs and placed on the Y haplotree.
Sure enough, a few days later, my McNeill cousin had a new haplogroup assignment.
Most people have at least one Private Variant, locations in which they do NOT match another tester. When several people have these same mutations, and they are high-quality reads, the Private Variant qualifies to be added to the haplotree as a SNP, a task performed at FamilyTreeDNA by Michael Sager.
If you ever have the opportunity to hear Michael speak, please do so. You can watch Michael’s presentation at Genetic Genealogy Ireland (GGI) titled “The Tree of Mankind,” on YouTube, here, compliments of Maurice Gleeson who coordinates GGI. Maurice has also written about the Gleeson Y DNA project analysis, here.
As a result of Cousin McNeill’s test, six new SNPs have been added to the Y haplotree, the tree of mankind. You can see our new haplogroup for our branch, BY18332, with an equivalent SNP, BY25938, along with three sibling branches to the left and right on the tree.

Big Y testing not only answers genealogical questions, it advances science by building out the tree of mankind too.
The surname of the men who share the same haplogroup, R-BY18332, meaning the named SNP furthest down the tree, are McCollum and Campbell. Not what I expected. I expected to find a McNeil who does match on at least some STR markers. This is exactly why the Big Y is so critical to define the tree structure, then use STR matches to flesh it out.
Taking the Big Y-700 test provided granularity between 6 matches, shown above, who were all initially assigned to the same branch of the tree, BY18350, but were subsequently divided into 4 separate branches. My McNiel cousin is no longer equally as distant from all 6 men. We now know that our McNiel line is genetically closer on the Y chromosome to Campbell and McCollum and further distant from Murphy, Scott, McMichael, and Glass.
Not All SNP Matches are STR Matches
Not all SNP matches are also STR matches. Some relationships are too far back in time. However, in this case, while each person on the BY18350 branches matches at some STR level, only the Campbell individual matches at all STR levels.
Remember that variants (mutations) are accumulating down both respective branches of the tree at the same time, meaning one per roughly every 100 years (if 100 is the average number we want to use) for both testers. A total of 30 variants or mutations difference, an average of 15 on each branch of the tree (McNiel and their match) would suggest a common ancestor about 1500 years ago, so each Big Y match should have a common ancestor 1500 years ago or closer. At least on average, in theory.
The Big Y test match threshold is 30 variants, so if there were any more mismatches with the Campbell male, they would not have been a Big Y match, even though they have the exact same haplogroup.
Having the same haplogroup means that their terminal SNP is identical, the SNP furthest down the tree today, at least until someone matches one of them on their Private Variants (if any remain unnamed) and a new terminal SNP is assigned to one or both of them.
Mutations, and when they happen, are truly a roll of the dice. This is why viewing all of your Big Y Block Tree matches is critical, even if they don’t show on your Big Y match list. One more variant and Campbell would have not been shown as a match, yet he is actually quite close, on the same branch, and matches on all STR panels as well.
SNPs Establish the Backbone Structure
I always view the block tree first to provide a branching tree structure, then incorporate STR matches into the equation. Both can equally as important to genealogy, but haplogroup assignment is the most accurate tool, regardless of whether the two individuals match on the Big Y test, especially if the haplogroups are relatively close.
Let’s work with the Block Tree.
The Block Tree

Clicking on the link to the Block Tree in the Big Y results immediately displays the tester’s branch on the tree, below.

click to enlarge
On the left side are SNP generation markers. Keep in mind that approximate SNP generations are marked every 5 generations. The most recent generations are based on the number of private variants that have not yet been assigned as branches on the tree. It’s possible that when they are assigned that they will be placed upstream someplace, meaning that placement will reduce the number of early branches and perhaps increase the number of older branches.
The common haplogroup of all of the branches shown here with the upper red arrow is R-BY3344, about 15 SNP generations ago. If you’re using 100 years per SNP generation, that’s about 1500 years. If you’re using 80 years, then 1200 years ago. Some people use even fewer years for calculations.
If some of the private variants in the closer branches disappear, then the common ancestral branch may shift to closer in time.
This tree will always be approximate because some branches can never be detected. They have disappeared entirely over time when no males exist to reproduce.
Conversely, subclades have been born since a common ancestor clade whose descendants haven’t yet tested. As more people test, more clades will be discovered.
Therefore, most recent common ancestor (MRCA) haplogroup ages can only be estimated, based on who has tested and what we know today. The tree branches also vary depending on whether testers have taken the Big Y-500 or the more sensitive Big Y-700, which detects more variants. The Y haplotree is a combination of both.
Big Y-500 results will not be as granular and potentially do not position test-takers as far down the tree as Big Y-700 results would if they upgraded. You’ll need to factor that into your analysis if you’re drawing genealogical conclusions based on these results, especially close results.
You’ll note that the direct path of descent is shown above with arrows from BY3344 through the first blue box with 5 equivalent SNPS, to the next white box, our branch, with two equivalent SNPs. Our McNeil ancestor, the McCollum tester, and the Campell tester have no unresolved private variants between them, which suggests they are probably closer in time than 10 generations back. You can see that the SNP generations are pushed “up” by the neighbor variants.
Because of the fact that private variants don’t occur on a clock cycle and occur in individual lines at an unsteady rate, we must use averages.
That means that when we look further “up” the tree, clicking generation by generation on the up arrow above BY3344, the SNP generations on the left side “adjust” based on what is beneath, and unseen at that level.
The Block Tree Adjusts
Note, in the example above, BY3344 is at SNP generation 15.
Next, I clicked one generation upstream, to R-S668.

click to enlarge
You can see that S668 is about 21 SNP generations upstream, and now BY3344 is listed as 20 generations, not 15. You can see our branch, BY3344, but you can no longer see subclades or our matches below that branch in this view.
You can, however, see two matches that descend through S668, brother branches to BY3344, red arrows at far right.
Clicking on the up arrow one more time shows us haplogroup S673, below, and the child branches. The three child branches on which the tester has matches are shown with red arrows.

click to enlarge
You’ll immediately notice that now S668 is shown at 19 SNP generations, not 20, and S673 is shown at 20. This SNP generation difference between views is a function of dealing with aggregated and averaged private variants on combined lines and causes the SNP generations to shift. This is also why I always say “about.”
As you continue to click up the tree, the shifting SNP generations continue, reminding us that we can’t truly see back in time. We can only achieve approximations, but those approximations improve as more people test, and more SNPs are named and placed in their proper places on the phylotree.
I love the Block Tree, although I wish I could see further side-to-side, allowing me to view all of the matches on one expanded tree so I can easily see their relationships to the tester, and each other.
Countries and Origins
In addition to displaying shared averaged autosomal origins of testers on a particular branch, if they have taken the Family Finder test and opted-in to sharing origins (ethnicity) results, you can also view the countries indicated by testers on that branch along with downstream branches of the tree.

click to enlarge
For example, the Countries tab for S673 is shown above. I can see matches on this branch with no downstream haplogroup currently assigned, as well as cumulative results from downstream branches.
Still, I need to be able to view this information in a more linear format.
The Block Tree and spreadsheet information beautifully augment the haplotree, so let’s take a look.
The Haplotree
On your Y DNA results page, click on the “Haplotree and SNPs” link.

click to enlarge
The Y haplotree will be displayed in pedigree style, quite familiar to genealogists. The SNP legend will be shown at the top of the display. In some cases, “presumed positive” results occur where coverage is lacking, back mutations or read errors are encountered. Presumed positive is based on positive SNPs further down the tree. In other words, that yellow SNP below must read positive or downstream ones wouldn’t.

click to enlarge
The tester’s branch is shown with the grey bar. To the right of the haplogroup-defining SNP are listed the branch and equivalent SNP names. At far right, we see the total equivalent SNPs along with three dots that display the Country Report. I wish the haplotree also showed my matches, or at least my matching surnames, allowing me to click through. It doesn’t, so I have to return to the Big Y page or STR Matches page, or both.
I’ve starred each branch through which my McNiell cousin descends. Sibling branches are shown in grey. As you’ll recall from the Block Tree, we do have matches on those sibling branches, shown side by side with our branch.
The small numbers to the right of the haplogroup names indicate the number of downstream branches. BY18350 has three, all displayed. But looking upstream a bit, we see that DF97 has 135 downstream branches. We also have matches on several of those branches. To show those branches, simply click on the haplogroup.
The challenge for me, with 119 McNeill matches, is that I want to see a combination of the block tree, my spreadsheet information, and the haplotree. The block tree shows the names, my spreadsheet tells me on which branches to look for those matches. Many aren’t easily visible on the block tree because they are downstream on sibling branches.
Here’s where you can find and view different pieces of information.
| Data and Sources |
STR Matches Page |
Big Y Matches Page |
Block Tree |
Haplogroups & SNPs Page |
| STR matches |
Yes |
No, but would like to see who matches at which STR levels |
If they have taken Big Y test, but doesn’t mean they match on Big Y matching |
No |
| SNP matches *1 |
Shows if STR match has common haplogroup, but not if tester matches on Big Y |
No, but would like to see who matches at which STR level |
Big Y matches and STR matches that aren’t Big Y matches are both shown |
No, but need this feature – see combined haplotree/ block tree |
| Other Haplogroup Branch Residents |
Yes, both estimated and tested |
No, use block tree or click through to profile card, would like to see haplogroup listed for Big Y matches |
Yes, both Big Y and STR tested, not estimated. Cannot tell if person is Big Y match or STR match, or both. |
No individuals, but would like that as part of countries report, see combined haplotree/block tree |
| Fully Expanded Phylotree |
No |
No |
Would like ability to see all branches with whom any Big Y or STR match resides at one time, even if it requires scrolling |
Yes, but no match information. Matches report could be added like on Block Tree. |
| Averaged Ethnicities if Have FF Test |
No |
No |
Yes, by haplogroup branch |
No |
| Countries |
Matches map STR only |
No, need Big Y matches map |
Yes |
Yes |
| Earliest Known Ancestor |
Yes |
No, but can click through to profile card |
No |
No |
| Customer Trees |
Yes |
No, need this link |
No |
No |
| Profile Card |
Yes, click through |
Yes, click through |
Yes, click through |
No match info on this page |
| Downloadable data |
By STR panel only, would like complete download with 1 click, also if Big Y or FF match |
Not available at all |
No |
No |
| Path to common haplogroup |
No |
No, but would like to see matches haplogroup and convergent haplogroup displayed |
No, would like the path to convergent haplogroup displayed as an option |
No, see combined match-block -haplotree in next section |
*1 – the best way to see the haplogroup of a Big Y match is to click on their name to view their profile card since haplogroup is not displayed on the Big Y match page. If you happen to also match on STRs, their haplogroup is shown there as well. You can also search for their name using the block tree search function to view their haplogroup.
Necessity being the mother of invention, I created a combined match/block tree/haplotree.
And I really, REALLY hope Family Tree DNA implements something like this because, trust me, this was NOT fun! However, now that it’s done, it is extremely useful. With fewer matches, it should be a breeze.
Here are the steps to create the combined reference tree.
Combo Match/Block/Haplotree
I used Snagit to grab screenshots of the various portions of the haplotree and typed the surnames of the matches in the location of our common convergent haplogroup, taken from the spreadsheet. I also added the SNP generations in red for that haplogroup, at far left, to get some idea of when that common ancestor occurred.

click to enlarge
This is, in essence, the end-goal of this exercise. There are a few steps to gather data.
Following the path of two matches (the tester and a specific match) you can find their common haplogroup. If your match is shown on the block tree in the same view with your branch, it’s easy to see your common convergent parent haplogroup. If you can’t see the common haplogroup, it’s takes a few extra steps by clicking up the block tree, as illustrated in an earlier section.
We need the ability to click on a match and have a tree display showing both paths to the common haplogroup.

I simulated this functionality in a spreadsheet with my McNiel cousin, a Riley match, and an Ocain match whose terminal SNP is the convergent SNP (M222) between Riley and McNiel. Of course, I’d also like to be able to click to see everyone on one chart on their appropriate branches.
Combining this information onto the haplotree, in the first image, below, M222, 4 men match my McNeill cousin – 2 who show M222 as their terminal SNP, and 2 downstream of M222 on a divergent branch that isn’t our direct branch. In other words, M222 is the convergence point for all 4 men plus my McNeill cousin.

click to enlarge
In the graphic below, you can see that M222 has a very large number of equivalent SNPs, which will likely become downstream haplogroups at some point in the future. However, today, these equivalent SNPs push M222 from 25 generations to 59. We’ll discuss how this meshes with known history in a minute.

click to enlarge
Two men, Ocain and Ransom, who have both taken the Big Y, whose terminal SNP is M222, match my McNiel cousin. If their common ancestor was actually 59 generations in the past, it’s very, very unlikely that they would match at all given the 30 mutation threshold.
On my reconstructed Match/Block/Haplotree, I included the estimated SNP generations as well. We are starting with the most distant haplogroups and working our way forward in time with the graphics, below.
Make no mistake, there are thousands more men who descend from M222 that have tested, but all of those men except 4 have more than 30 mutations total, so they are not shown as Big Y matches, and they are not shown individually on the Block Tree because they neither match on the Big Y or STR tests. However, there is a way to view information for non-matching men who test positive for M222.

click to enlarge
Looking at the Block Tree for M222, many STR match men took a SNP test only to confirm M222, so they would be shown positive for the M222 SNP on STR results and, therefore, in the detailed view of M222 on the Block tree.
Haplogroup information about men who took the M222 test and whom the tester doesn’t match at all are shown here as well in the country and branch totals for R-M222. Their names aren’t displayed because they don’t match the tester on either type of Y DNA test.
Back to constructing my combined tree, I’ve left S658 in both images, above and below, as an overlap placeholder, as we move further down, or towards current, on the haplotree.

click to enlarge
Note that BY18350, above, is also an overlap connecting below.
You’ll recall that as a result of the Big Y test, BY18350 was split and now has three child branches plus one person whose terminal SNP is BY18350. All of the men shown below were on one branch until Big Y results revealed that BY18350 needed to be split, with multiple new haplogroups added to the tree.

click to enlarge
Using this combination of tools, it’s straightforward for me to see now that our McNiel line is closest to the Campbell tester from Scotland according to the Big Y test + STRs.
Equal according to the Big Y test, but slightly more distant, according to STR matching, is McCollum. The next closest would be sibling branches. Then in the parent group of the other three, BY18350, we find Glass from Scotland.
In BY18350 and subgroups, we find several Scotland locations and one Northern Ireland, which was likely from Scotland initially, given the surname and Ulster Plantation era.
The next upstream parent haplogroup is BY3344, which looks to be weighted towards ancestors from Scotland, shown on the country card, below.

click to enlarge
This suggests that the origins of the McNiel line was, perhaps, in Scotland, but it doesn’t tell us whether or not George and presumably, Thomas, immigrated from Ireland or Scotland.
This combined tree, with SNPs, surnames from Big Y matches, along with Country information, allows me to see who is really more closely related and who is further away.
What I didn’t do, and probably should, is to add in all of the STR matches who have taken the Big Y test, shown on their convergent branch – but that’s just beyond the scope of time I’m willing to invest, at least for now, given that hundreds of STR matches have taken the Big Y test, and the work of building the combined tree is all manual today.
For those reading this article without access to the Y phylogenetic tree, there’s a public version of the Y and mitochondrial phylotrees available, here.
What About Those McNiels?
No other known McNiel descendants from either Thomas or George have taken the Big Y test, so I didn’t expect any to match, but I am interested in other men by similar surnames. Does ANY other McNiel have a Big Y match?
As it turns out, there are two, plus one STR match who took a Big Y test, but is not a Big Y match.
However, as you can see on the combined match/block/haplotree, above, the closest other Big Y-matching McNeil male is found at about 19 SNP generations, or roughly 1900 years ago. Even if you remove some of the variants in the lower generations that are based on an average number of individual variants, you’re still about 1200 years in the past. It’s extremely doubtful that any surname would survive in both lines from the year 800 or so.
That McNeil tester’s ancestor was born in 1747 in Tranent, Scotland.
The second Big Y-matching person is an O’Neil, a few branches further up in the tree.
The convergent SNP of the two branches, meaning O’Neil and McNeill are at approximately the 21 generation level. The O’Neil man’s Neill ancestor is found in 1843 in Cookestown, County Tyrone, Ireland.

I created a spreadsheet showing convergent lines:
- The McNeill man with haplogroup A4697 (ancestor Tranent, Scotland) is clearly closest genetically.
- O’Neill BY91591, who is brother clades with Neel and Neal, all Irish, is another Big Y match.
- The McNeill man with haplogroup FT91182 is an STR match, but not a Big Y match.
The convergent haplogroup of all of these men is DF105 at about the 22 SNP generation marker.
STRs
Let’s turn back to STR tests, with results that produce matches closer in time.
Searching my STR download spreadsheet for similar surnames, I discovered several surname matches, mining the Earliest Known Ancestor information, profiles and trees produced data as follows:
| Ancestor |
STR Match Level |
Location |
| George Charles Neil |
12, 25, match on Big Y A4697 |
1747-1814 Tranent, Scotland |
| Hugh McNeil |
25 (tested at 67) |
Born 1800 Country Antrim, Northern Ireland |
| Duncan McNeill |
12 (tested at 111) |
Married 1789, Argyllshire, Scotland |
| William McNeill |
12, 25 (tested at 37) |
Blackbraes, Stirlingshire, Scotland |
| William McNiel |
25 (tested at 67) |
Born 1832 Scotland |
| Patrick McNiel |
25 (tested at 111) |
Trien East, County Roscommon, Ireland |
| Daniel McNeill |
25 (tested at 67) |
Born 1764 Londonderry, Northern Ireland |
| McNeil |
12 (tested at 67) |
1800 Ireland |
| McNeill (2 matches) |
25 (tested Big Y- SNP FT91182) |
1810, Antrim, Northern Ireland |
| Neal |
25 – (tested Big Y, SNP BY146184) |
Antrim, Northern Ireland |
| Neel (2 matches) |
67 (tested at 111, and Big Y) |
1750 Ireland, Northern Ireland |
Our best clue that includes a Big Y and STR match is a descendant of George Charles Neil born in Tranent, Scotland, in 1747.
Perhaps our second-best clue comes in the form of a 111 marker match to a descendant of one Thomas McNeil who appears in records as early as 1753 and died in 1761 In Rombout Precinct, Dutchess County, NY where his son John was born. This line and another match at a lower level both reportedly track back to early New Hampshire in the 1600s.
The MacNeil DNA Project tells us the following:
Participant 106370 descends from Isaiah McNeil b. 14 May 1786 Schaghticoke, Rensselaer Co. NY and d. 28 Aug 1855 Poughkeepsie, Dutchess Co., NY, who married Alida VanSchoonhoven.
Isaiah’s parents were John McNeal, baptized 21 Jun 1761 Rombout, Dutchess Co., NY, d. 15 Feb 1820 Stillwater, Saratoga Co., NY and Helena Van De Bogart.
John’s parents were Thomas McNeal, b.c. 1725, d. 14 Aug 1761 NY and Rachel Haff.
Thomas’s parents were John McNeal Jr., b. around 1700, d. 1762 Wallkill, Orange Co., NY (now Ulster Co. formed 1683) and Martha Borland.
John’s parents were John McNeal Sr. and ? From. It appears that John Sr. and his family were this participant’s first generation of Americans.
Searching this line on Ancestry, I discovered additional information that, if accurate, may be relevant. This lineage, if correct, and it may not be, possibly reaching back to Edinburgh, Scotland. While the information gathered from Ancestry trees is certainly not compelling in and of itself, it provides a place to begin research.
Unfortunately, based on matches shown on the MacNeil DNA Project public page, STR marker mutations for kits 30279, B78471 and 417040 when compared to others don’t aid in clustering or indicating which men might be related to this group more closely than others using line-marker mutations.
Matches Map
Let’s take a look at what the STR Matches Map tells us.

This 67 marker Matches Map shows the locations of the earliest known ancestors of STR matches who have entered location information.


My McNeill cousin’s closest matches are scattered with no clear cluster pattern.
Unfortunately, there is no corresponding map for Big Y matches.
SNP Map
The SNP map provided under the Y DNA results allows testers to view the locations where specific haplogroups are found.

The SNP map marks an area where at least two or more people have claimed their most distant known ancestor to be. The cluster size is the maximum amount of miles between people that is allowed in order for a marker indicating a cluster at a location to appear. So for example, the sample size is at least 2 people who have tested, and listed their most distant known ancestor, the cluster is the radius those two people can be found in. So, if you have 10 red dots, that means in 1000 miles there are 10 clusters of at least two people for that particular SNP. Note that these locations do NOT include people who have tested positive for downstream locations, although it does include people who have taken individual SNP tests.
Working my way from the McNiel haplogroup backward in time on the SNP map, neither BY18332 nor BY18350 have enough people who’ve tested, or they didn’t provide a location.
Moving to the next haplogroup up the tree, two clusters are formed for BY3344, shown below.

S668, below.

It’s interesting that one cluster includes Glasgow.
S673, below.

DF85, below:

DF105 below:

M222, below:

For R-M222, I’ve cropped the locations beyond Ireland and Scotland. Clearly, RM222 is the most prevalent in Ireland, followed by Scotland. Wherever M222 originated, it has saturated Ireland and spread widely in Scotland as well.
R-M222
R-M222, the SNP initially thought to indicate Niall of the 9 Hostages, occurred roughly 25-59 SNP generations in the past. If this age is even remotely accurate, averaging by 80 years per generation often utilized for Big Y results, produces an age of 2000 – 4720 years. I find it extremely difficult to believe any semblance of a surname survived that long. Even if you reduce the time in the past to the historical narrative, roughly the year 400, 1600 years, I still have a difficult time believing the McNiel surname is a result of being a descendant of Niall of the 9 Hostages directly, although oral history does have staying power, especially in a clan setting where clan membership confers an advantage.
Surname or not, clearly, our line along with the others whom we match on the Big Y do descend from a prolific common ancestor. It’s very unlikely that the mutation occurred in Niall’s generation, and much more likely that other men carried M222 and shared a common ancestor with Niall at some point in the distant past.
McNiel Conclusion – Is There One?
If I had two McNiel wishes, they would be:
- Finding records someplace in Virginia that connect George and presumably brothers Thomas and John to their parents.
- A McNiel male from wherever our McNiel line originated becoming inspired to Y DNA test. Finding a male from the homeland might point the way to records in which I could potentially find baptismal records for George about 1720 and Thomas about 1724, along with possibly John, if he existed.
I remain hopeful for a McNiel from Edinburgh, or perhaps Glasgow.
I feel reasonably confident that our line originated genetically in Scotland. That likely precludes Niall of the 9 Hostages as a direct ancestor, but perhaps not. Certainly, one of his descendants could have crossed the channel to Scotland. Or, perhaps, our common ancestor is further back in time. Based on the maps, it’s clear that M222 saturates Ireland and is found widely in Scotland as well.
A great deal depends on the actual age of M222 and where it originated. Certainly, Niall had ancestors too, and the Ui Neill dynasty reaches further back, genetically, than their recorded history in Ireland. Given the density of M222 and spread, it’s very likely that M222 did, in fact, originate in Ireland or, alternatively, very early in Scotland and proliferated in Ireland.
If the Ui Neill dynasty was represented in the persona of the High King, Niall of the 9 Hostages, 1600 years ago, his M222 ancestors were clearly inhabiting Ireland earlier.
We may not be descended from Niall personally, but we are assuredly related to him, sharing a common ancestor sometime back in the prehistory of Ireland and Scotland. That man would sire most of the Irish men today and clearly, many Scots as well.
Our ancestors, whoever they were, were indeed in Ireland millennia ago. R-M222, our ancestor, was the ancestor of the Ui Neill dynasty and of our own Reverend George McNiel.
Our ancestors may have been at Knowth and New Grange, and yes, perhaps even at Tara.

Someplace in the mists of history, one man made a different choice, perhaps paddling across the channel, never to return, resulting in M222 descendants being found in Scotland. His descendants include our McNeil ancestors, who still slumber someplace, awaiting discovery.
_____________________________________________________________
Disclosure
I receive a small contribution when you click on some of the links to vendors in my articles. This does NOT increase the price you pay but helps me to keep the lights on and this informational blog free for everyone. Please click on the links in the articles or to the vendors below if you are purchasing products or DNA testing.
Thank you so much.
DNA Purchases and Free Transfers
Genealogy Products and Services
Genealogy Research
Like this:
Like Loading...




 You’ll notice that each parent carries more of each color DNA than they pass on to their own children, so different children receive different pieces of their parents’ DNA, and varying percentages of their grandparents’ DNA.
You’ll notice that each parent carries more of each color DNA than they pass on to their own children, so different children receive different pieces of their parents’ DNA, and varying percentages of their grandparents’ DNA.![]()
![]()

![]()