Genetic Affairs has created a new version of AutoKinship at GEDmatch. The new AutoKinship report adds new features, allows for more kits to be included in the analysis, and integrates multiple reports together:
- AutoCluster – the autoclusters we all know and love
- AutoSegment – clusters based on segments
- AutoTree – reconstructed tree based on GEDCOM files of you and your matches, even if you don’t have a tree
- AutoKinship – the original AutoKinship report provided genetic trees. The new AutoKinship report includes AutoTree, combines both, and adds features called AutoKinship Tree. (Trust me on this one – you’ll see in a minute!)
- Matches
- Common Ancestors with your ancestors
- Common Ancestors between matches, even if they don’t match your tree
- Common Locations
Maybe the best news is that some reports provide automatic triangulation because, at GEDmatch, it’s possible to not only see how you match multiple people, but also if those people match each other on that same segment. Of course, triangulation requires three-way matching in addition to the identification of common ancestors which is part of what AutoKinship provides, in multiple ways.
Let’s step through the included reports and features one at a time, using my clusters as an example.
Order Your Report
As a Tier 1 GEDmatch customer, sign in, select AutoKinship and order your report.
Note that there are now two clustering settings, the default setting and one that will provide more dense clusters. The last setting is the default setting for AutoKinship, since it has been shown to produce better AutoKinship results.
You can also select the number of kits to consider. Since this tool is free with a GEDmatch Tier 1 subscription, you can start small and rerun if you wish, as often as you wish.
Currently, a maximum of 500 matches can be included, but that will be increased to 1000 in the future. Your top 500 matches will be included that fall within the cM matching parameters specified.
I’m leaving this at the maximum 400 cM threshold, so every match below that is included. I generally leave this default threshold because otherwise my closest matches will be in a huge number of clusters which may cause processing issues.
For a special use case where you will want to increase the cM threshold, see the Special Use Cases section near the end of this article.
You can select a low number of matches, like 25 or 50 which is particularly useful if you want to examine the closest matches of a kit without a tree.
Keep in mind that there is currently a maximum processing time of 10 minutes allowed per report. This means that if you have large clusters, which are the last ones processed, you may not have AutoKinship results for those clusters.
This also means that if you select a high cM threshold and include all 500 allowable matches, you will receive the report but the AutoKinship results may not be complete.
When finished, your report will be delivered to you as a download link with an attached zipped file which you will need to save someplace where you can find it.
Unzip
If you’re a PC user, you’ll need to unzip or extract the files before you can use the files. You’ll see the zipper on the file.
If you don’t extract the contents, you can click on the file to open which will display a list of the files, so it looks like the files are extracted, but they aren’t.
You can see that the file is still zipped.
You can click on the html file which will display the AutoCluster correctly too, but when you click on any other link within that file, you’ll receive this error message if the file is still zipped.
If this happens to you, it means the file is still zipped. Close the files you have open, right click on the yellow zipped file folder and “extract all.”
Then click on the HTML link again and everything should work.
Ok, on to the fun part – the tools.
Tools
I’ve written about most of these tools individually before, except for the new combinations of course. I’ve put all of the Genetic Affairs Tools, Instructions and Resources in one article that you can find here.
I recommend that you take a look to be sure you’re using each tool to its greatest advantage.
AutoCluster
Click on the html file and watch your AutoCluster fly into place. I always, always love this part.
The first thing I noticed about my AutoCluster at GEDmatch is that it’s HUGE! I have a total of 144 clusters and that’s just amazing!
Information about the cluster file, including the number of matches, maximum and minimum cM used for the report, and minimum cluster size appears beneath your cluster chart.
22 people met the criteria but didn’t have other matches that did, so they are listed for my review, but not included in the cluster chart.
At first glance, the clusters look small, but don’t despair, they really aren’t.
My clusters only look small because the tool was VERY successful, and I have many matches in my clusters. The chart has to be scaled to be able to display on a computer monitor.
New Layout
Genetic Affairs has introduced a new layout for the various included tools.
Each section opens to provide a brief description of the tool and what is occurring. This new tool includes four previous tools plus a new one, AutoCluster Tree, as follows:
AutoCluster
AutoCluster first organizes your DNA matches into shared match clusters that likely represent branches of your family. Everyone in a cluster will likely be on the same ancestral line, although the MRCA between any of the matches and between you and any match may vary. The generational level of the clusters may vary as well. One may be your paternal grandmother’s branch, another may be your paternal grandfather’s father’s branch.
AutoSegment
AutoSegment organizes your matches based on triangulating segments. AutoSegment employs the positional information of segments (chromosome and start and stop position) to identify overlapping segments in order to link DNA matches. In addition, triangulated data is used to collaborate these links. Using the user defined minimum overlap of a DNA segment we perform a clustering of overlapping DNA segments to identify segment clusters. The overlap is calculated in centimorgans using human genetic recombination maps. Another aspect of overlapping segments is the fact that some regions of our genome seem to have more matches as compared to the other regions. These so-called pile-up areas can influence the clustering. The removal of known pile-up regions based on the paper of Li et al 2014 is optional and is not performed for this analysis However, a pileup report is provided that allows you to examine your genome for pileup regions.
AutoTree
By comparing the tree of the tested person and the trees from the members of a certain cluster, we can identify ancestors that are common amongst those trees. First, we collect the surnames that are present in the trees and create a network using the similarity between surnames. Next, we perform a clustering on this network to identify clusters of similar surnames. A similar clustering is performed based on a network using the first names of members of each surname cluster. Our last clustering uses the birth and death years of members of a cluster to find similar persons. As a consequence, initially large clusters (based on the surnames) are divided up into smaller clusters using the first name and birth/death year clustering.
AutoKinship
AutoKinship automatically predicts family trees based on the amount of DNA your DNA matches share with you and each other. Note that AutoKinship does not require any known genealogical trees from your DNA matches. Instead, AutoKinship looks at the predicted relationships between your DNA matches, and calculates many different paths you could all be related to each other. The probabilities used by this AutoKinship analysis are based on simulated data for GEDmatch matches and are kindly provided by Brit Nicholson (methodology described here). Based on the shared cM data between shared matches, we create different trees based on the putative relationships. We then use the probabilities to test every scenario which are then ranked.
AutoKinship Tree
Predicted trees from the AutoTree analysis are based on genealogical trees shared by the DNA matches and, if available, shared by the tested person. The relationships between DNA matches based on their common ancestors as provided AutoTree are used to perform an AutoKinship analysis and are overlayed on the predicted AutoKinship tree.
AutoKinship Tree is New
AutoKinship Tree is the new feature that combines the features of both AutoTree and AutoKinship. You receive:
- Common ancestors between you and your matches
- Trees of people who don’t share your common ancestors but share ancestors with each other
- Combined with relationship predictions and
- A segment analysis
Of course, the relative success of the tree tools depends upon how many people have uploaded GEDCOM files.
Big hint, if you haven’t uploaded your family tree, do so now. If you are an adoptee or searching for a parent and don’t know who your ancestors are, AutoKinship Tree does its best without your tree information, and you will still benefit from the trees of others combined with predicted relationships based on DNA.
It’s easier to show you than to tell you, so let’s step through my results one section at a time.
I’m going to be using cluster 5 which has 32 members and cluster 136 which has 8 members. Ironically, cluster 136 is a much more useful cluster, with 8 good matches, than cluster 5 which includes 32 people.
Results of the AutoKinship Analyses
As you scroll down your results, you’ll see a grid beneath the Explanation area.
It’s easy to see which cluster received results for each tool. My cluster 5 has results in each category, along with surnames. (Notice that you can search for surnames which displays only the clusters that contain that surname.)
I can click on each icon to see what’s there waiting for me.
Additionally, you can click at the top on the blue middle “here” for an overview of all common ancestors. Who can resist that, right?
Click on the ancestor’s name or the tree link to view more information.
You can also view common locations too by clicking on the blue “here” at far right. A location, all by itself, is a HUGE hint.
Clicking on the tree link shows you the tree of the tester with ancestors at that location. I had several others from North Carolina, generally, and other locations specifically. Let’s take a look at a few examples.
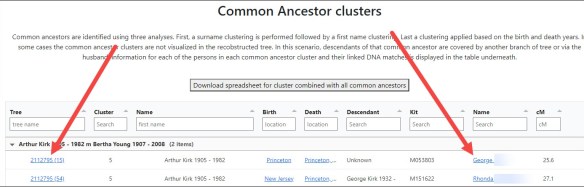
Common Ancestor Clusters
Click on the first blue link to view all common ancestors.
Common Ancestor Clusters summarize all of the clusters by ancestor. In other words, if any of your matches have ancestors in common in their tree, they are listed here.
These clusters include NOT just the people who share ancestors in a tree with you, but who also share known ancestors with each other BUT NOT YOU. That may be incredibly important when you are trying to identify your ancestors – as in brick walls. Your ancestors may be their ancestors too, or your common segments might lead to your common ancestors if you complete their tree.
There are other important hints too.
In my case, above, Jacob Lentz is my known ancestor.
However, Sarah Barron is not my ancestor, nor is John Vincent Dodson. They are the descendants of my Dodson ancestor though. I recognized that surname and those people. In other instances, recognizing a common geography may be your clue for figuring out how you connect.
In the cluster column at left, you can see the cluster number in which these people are found.
Common Locations Table
Clicking on the second link provides a Common Location Table
Some locations are general, like a state, and others are town, county or even village names. Whatever people have included in their GEDCOM files that can be connected.
Looking at this first entry, I recognize some of the ancestral surnames of Karen’s ancestors. The fact that we are found in the same cluster and share DNA indicates a common ancestor someplace.
Check for this same person in additional locations, then, look at their tree.
Ok, back to the AutoKinship Analysis Table and Cluster 136.
Cluster 136
I’m going to use Cluster 136 as an example because this cluster has generated great reports using all of the tools, indicated by the icon under each column heading. Some clusters won’t have enough information for everything so the tools generate as much as possible.
Scrolling down to Cluster 136 in the AutoCluster Information report, just beneath the list of clusters, I can see my 8 matches in that cluster.
Of course, I can click on the links for specific information, or contact them via email. At the end of this article in the “Tell Me Everything” section, I’ll provide a way to retrieve as much information as possible about any one match. For now, let’s move to the AutoTree.
Cluster 136 AutoTree
Clicking on the icon under AutoTree shows me how two of the matches in this cluster are related to each other and myself.
Note that the centimorgan badges listed refer to the number of cM that I share with each of these people, not how much they share with each other.
Click on any of the people to see additional information.
When I click on J Lentz m F Moselman, a popup box shows me how this couple is related to me and my matches.
Of course, you can also view the Y DNA or mitochondrial DNA haplogroups if the testers have provided that information when they set up their GEDmatch profile information.
 Just click on the little icons.
Just click on the little icons.
If the testers have not provided that information, you can always check at FamilyTreeDNA or 23andMe, if they have tested at either of those vendors, to view their haplogroup information.
Today, GEDmatch kit numbers are assigned randomly, but in the early days, before Genesis, the leading letter of A meant AncestryDNA, F or T for FamilyTreeDNA, M for 23andMe and H for MyHeritage. If the kit number is something else, perform a one-to-one or a one-to-many report which will display the source of their DNA file.
The small number, 136 in this case, beside the cM number indicates the cluster or clusters that these people are members of. Some people are members of multiple clusters
Let’s see what’s next.
Cluster 136 Common Ancestors
Clicking on the Ancestors icon provides a report that shows all of the Ancestor Clusters in cluster 136.
The difference between this ancestor chart and the larger chart is that this only shows ancestors for cluster 136, while the larger chart shows ancestors for the entire AutoCluster report.
Cluster 136 Locations
All of the locations shown are included in trees of people who cluster together in cluster 136. Of course, this does NOT mean that these locations are all relevant to cluster 136. However, finding my own tree listed might provide an important clue.
Using the location tool, I discover 5 separate location clusters. This location cluster includes me with each tester’s ancestors who are found in Montgomery County, Ohio.
The difference between this chart for cluster 136 only and the larger location chart is that every location in this chart is relevant for people who all cluster together meaning we all share some ancestral line.
Viewing the trees of other people in the cluster may suggest ancestors or locations that are essential for breaking down brick walls.
Cluster 136 AutoKinship
Clicking on the anchor in the AutoKinship column provides a genetically reconstructed tree based on how closely each of the people match me, and each other. Clearly, in order to be able to provide this prediction, information about how your matches also match each other, or don’t, is required.
Again, the cM amount shown is the cM match with me, not with each other. However, if you click on a match, a popup will be shown that shows the shared cM between that person and the other matches as well as the relationship prediction between them in this tree
So, Bill matches David with a total of 354.3 cM and they are positioned as first cousins once removed in this tree. The probability of the match being a 1C1R (first cousin once removed) is 64.9%, meaning of course that other relationships are possible.
Note that Bill and David ALSO share a segment with me in autosegment cluster 185, on chromosome 3.
It’s important to note that while 136 is the autocluster number, meaning that colored block on the report, WITHIN clusters, autosegment clusters are formed and numbered.
Each autosegment cluster receives its own number and the numbers are for the entire report. You will have more autosegment clusters than autoclusters, because at least some of the colorful autoclusters will contain more than one segment cluster.
Remember, autoclusters are those colorful boxes of matches that fly into place. Autosegment clusters are the matching triangulated clusters on chromosomes and they are represented by the blue bars, shown below.
AutoCluster 136 contains 5 different autosegment clusters, but Bill is only included in one of those autosegment clusters.
You’ll notice that there are some people, like Robin at the bottom, who do match some other people in the cluster, but either not enough people, or not enough overlapping DNA to be included as an autocluster member.
The small colored chromosomes with numbers, boxed in red, indicate the chromosome on which this person matches me.
If you click on that chromosome icon, you’ll see a popup detailing everyone who matches me on that segment.
Note that in some cases a member of a segment cluster, like Robin, did not make it in the AutoCluster cluster. You can spot these occurrences by scrolling down and looking at the cluster column which will then be empty for that particular match.
Reconstructed AutoKinship Trees in Most Likely Order
Scrolling down the page, next we see that we have multiple possible trees to view. We are shown the most likely tree first.
Tree likelihood is constructed based on the combined probability of my matching cM to an individual plus their likely relationship to each other based on the amount of DNA they share with each other as well.
In my case, all of the first 8 trees are equally as likely to be accurate, based on autosomal genetic relationships only. The ninth tree is only very slightly less likely to be accurate.
The X chromosome is not utilized separately in this analysis, nor are Y or mitochondrial DNA haplogroups if provided.
DNA Relationship Matrix
Continuing to scroll down, we next see the DNA matrix that shows relationships for cluster 5 in a grid format. Click on “Download Relationship Matrix” to view in a spreadsheet.
Keep scrolling for the next view which is the Individual Segment Cluster Information
Individual Segment Cluster Information
Remember that we are still focused on only one cluster – in this case, cluster 136. Each cluster contains people who all match at least some subset of other people in the cluster. Some people will match each other and the tested person on the same chromosome segment, and some won’t. What we generally see within clusters are “subclusters” of people who match each other on different chromosomes and segments. Also, some matches from cluster 136 might match other people but those matches might not be a member of cluster 136.
In autocluster 136, I have 14 DNA segments that converge into 5 segment clusters with my matches. Here’s segment cluster 185 that consists of two people in addition to me. Note that for individuals to be included in these segment clusters at GEDmatch, they must triangulate with people in the same segment cluster.
From left to right, we see the following information:
- AutoCluster number 136, shown below
- Segment cluster 185. This is a segment cluster within autocluster 136.

- Segment cluster 185 occurs on chromosome 3, between the designated start and stop locations.
- The segment representation shows the overlapping portions of the two matches, to me. You can easily see that they overlap almost exactly with each other as well.
- The SNP count is shown, followed by the name and cM count.
Cluster 136 AutoKinship Tree
The AutoKinship Tree column is different from the AutoKinship column in one fundamental way. The new AutoKinship Tree feature combines the genealogical AutoTree and the genetic AutoKinship output together in one report.
You can see that the “prior” genealogical tree information that one of my matches also descends from Jacob Lentz (and wife, if you click further) has now been included. The matches without trees have been reconstructed around the known genealogy based on how they match me and each other.
I was already aware of how I’m related to Bill, David, *C and *R, but I don’t know how I am related to these other people. Based on their kit identifier, I can go to the vendor where they tested and utilize tools there, and I can check to see if they have uploaded their DNA files elsewhere to discover additional records information or critical matches. Now at least I know where in the tree to search.
Cluster 136 AutoSegment
Clicking on AutoSegment provides you with segment information. Each cluster is painted on your chromosomes.
By hovering over the darkly colored segments, which are segment clusters, you can view who you match, although to view multiple matches, continue scrolling.
In the next section, you’ll see the two segment clusters contained wholly within cluster 136.
Following that is the same information for segment clusters partially linked to cluster 136, but not contained wholly within 136.
Bonus – Tell Me Everything – Individual Match Clusters
We’ve focused specifically on the AutoKinship tools, but if you’re interested in “everything” about one specific match, you can approach things from that perspective too. I often look at a cluster, then focus on individuals, beginning with those I can identify which focuses my search.
If you click on any person in your match list, you’ll receive a report focusing on that person in your autocluster.
Let’s use cousin Bill as an example. I know how he’s related to me.
You can choose to display your chosen cluster by:
- Cluster
- Number of shared matches
- Shared cM with the tester
- Name
I would suggest experimenting with all of the options and see which one displays information that is most useful to the question you’re trying to answer.
Beneath the cluster for Bill, you’ll see the relevant information about the cluster itself. Bill has cluster matches on two different chromosomes.
The AutoCluster Cluster member Information report shows you how much DNA each cluster member shares with the tested person, which is me, and with each other cluster member. It’s easy to see at a glance who Bill is most closely related to by the number of cMs shared.
Only one of Bill’s chromosomes, #3, is included in clusters, but this tells me immediately that this/these segments on chromosome 3 triangulate between me, Bill, and at least one other person.
Segments shown in orange (chromosome 22) match me, but are not included in a cluster.
Special Use Cases – Unknown People
For adoptees and people trying to figure out how they are related to closer relatives, especially those without a tree, this new combined AutoKinship tool is wonderful.
400 cM is the upper default limit when running the report, meaning that close family members will not be included because they would be included in many clusters. However, you can make a different selection. If you’re trying to determine how several closely related people intersect, select a high threshold to include everyone.
Select a lower number of matches, like 25 or 50.
In this example, ‘no limit” was selected as the upper total match threshold and 25 closest matches.
AutoKinship then constructs a genetic tree and tells you which trees are possible and most likely. If some people do have trees, that common ancestor information would be included as well.
Note that when matches occur over the 400 cM threshold, there will be too many common chromosome matches so the chromosome numbers are omitted. Just check the other reports.
This tool would have helped a great deal with a recent close match who didn’t know how they are related to my family.
You can see this methodology in action and judge its accuracy by reconstructing your own family, assuming some of your known family members have uploaded to GEDmatch. Try it out.
It’s a Lot!
I know there’s a lot here to absorb, but take your time and refer back to this article as needed.
This flexible new tool combines DNA matching, genealogy trees, genetic trees, locations, autoclusters, a chromosome browser, and triangulation. It took me a few passes and working with different clusters to understand and absorb the information that is being provided.
For people who don’t know who their parents or close relatives are, these tools are amazing. Not only can they determine who they are related to, and who is related to each other, but with the use of trees, they can view common ancestors which provides possible ancestors for them too.
For people painting their triangulated segments at DNAPainter, AutoKinship provides triangulation groups that can be automatically painted using the Cluster Auto Painter, here, plus helps to identify that common ancestor. You can read more about DNAPainter, here.
For people seeking to break down brick walls, AutoKinship Tree provides assistance by providing tree matching between your matches for common ancestors NOT IN YOUR TREE, but that ARE in theirs. Your brick walls are clearly not (yet) identified in your tree, although that’s our fervent hope, right?
Even if your matches’ trees don’t go far enough back, as a genealogist, you can extend those trees further to hopefully reveal a previously unknown common ancestor.
The Best Things You Can Do
Aside from DNA testing, the three best things you can do to help yourself, and your clusters are:
- Upload your GEDCOM file, complete with locations, so you have readily available trees. Ask your matches to do so as well. Trees help you and others too.
- Encourage people you match at Ancestry who provides no chromosome segment information or chromosome browser to upload a copy of their DNA files and tree.
- Test your family members and cousins, and encourage them to upload their DNA and their trees. Offer to assist them. You can find step-by-step download/upload instructions here.
Have fun!
______________________________________________________________
Sign Up Now – It’s Free!
If you enjoyed this article, subscribe to DNAeXplain for free to automatically receive new articles by email each week.
Here’s the link. Just look for the little grey “follow” button on the right-hand side on your computer screen below the black title bar, enter your e-mail address, and you’re good to go!
In case you were wondering, I never have nor ever will share or use your e-mail outside of the intended purpose.
Share the Love
You can always forward these articles to friends or share by posting links on social media. Who do you know that might be interested?
_____________________________________________________________
Follow DNAexplain on Facebook, here or follow me on Twitter, here.
Share the Love!
You’re always welcome to forward articles or links to friends and share on social media.
If you haven’t already subscribed (it’s free,) you can receive an email whenever I publish by clicking the “follow” button on the main blog page, here.
You Can Help Keep This Blog Free
I receive a small contribution when you click on some of the links to vendors in my articles. This does NOT increase the price you pay but helps me to keep the lights on and this informational blog free for everyone. Please click on the links in the articles or to the vendors below if you are purchasing products or DNA testing.
Thank you so much.
DNA Purchases and Free Uploads
- FamilyTreeDNA – Y, mitochondrial and autosomal DNA testing
- MyHeritage DNA – Autosomal DNA test
- MyHeritage FREE DNA file upload – Upload your DNA file from other vendors free
- AncestryDNA – Autosomal DNA test
- 23andMe Ancestry – Autosomal DNA only, no Health
- 23andMe Ancestry Plus Health
Genealogy Products and Services
- MyHeritage FREE Tree Builder – Genealogy software for your computer
- MyHeritage Subscription with Free Trial
- Legacy Family Tree Webinars – Genealogy and DNA classes, subscription-based, some free
- Legacy Family Tree Software – Genealogy software for your computer
- Newspapers.com – Search newspapers for your ancestors
- NewspaperArchive – Search different newspapers for your ancestors
My Book
- DNA for Native American Genealogy – by Roberta Estes, for those ordering within the United States
- DNA for Native American Genealogy – for those ordering outside the US
Genealogy Books
- Genealogical.com – Lots of wonderful genealogy research books
Genealogy Research
- Legacy Tree Genealogists – Professional genealogy research
Discover more from DNAeXplained - Genetic Genealogy
Subscribe to get the latest posts sent to your email.

















































Hi,
You note that “Currently, a maximum of 500 matches can be included, but that will be increased to 1000 in the future. Your top 500 matches will be included that fall within the cM matching parameters specified.”
Are the 500 matches enough to pull in matches that are around 20cM? I see you are showing some.
It depends on your situation. If you don’t have a lot of close ones, or you drop the upper threshold. Try it out.
I cannot understand how to use this tool. Some simple way should be used. Too complicated!
The easy way is to just run it, accept all the defaults, and click on each option to see what is there.
I just keep on getting message that says my file cannot be found – what should i do??
You haven’t saved the file and unzipped it. Save it, right click on the folder for and select “extract all.”
Never could understand all this way back when gen match came out years ago.
Any insights into using this tool for those of us whose ancestors came from endogamous villages?
I don’t. I do deal with some endogamy but not comparable to others. I would expect you’d see a lot of grey cells where people are in multiple clusters. Let me know what you are seeing.
Both times I’ve run this, I get only one cluster with everyone except the handful of people who “don’t have any shared matches” (even though I know I have shared matches with some of them.)
First time I ran it with the default settings, the next time I changed the upper cM threshold to 250cMs just to try something different.
Also, the clusters had no results under the Auto Tree or Ancestors columns.
When I did the stand alone auto cluster tool, I have 26 clusters ranging from 2-7 people each. (As a side note, I only have 3 matches above 100, one of them only just.)
Any suggestions on settings to use that will give me results that would be more useful than everyone in one cluster?
Jennifer
I don’t know. I’ll ask the designer to answer this comment or you can ask in the Genetic Affairs Facebook group.
hi Jennifer, here EJ from Genetic Affairs. Perhaps the new “dense clustering” setting is not working for your kit. Could you change it to the default and see what happens. Did you also try to select the top 500 matches? If it still fails, contact us at https://members.geneticaffairs.com/contactus
Well, changing out of “dense clusters” and the top 500 matches, I at least have 6 clusters now, though 414 matches are in one and the other 5 have 2-6 each. 2 of the 6 clusters have zero results in the analyses table. (one of which has 2 of the very few matches I actually know my relationship with.)
I guess I will just continue to play around with various settings to see what comes of it and will reach out at geneticaffairs if it’s not producing more helpful results.
Thanks!
Jennifer
I’m looking for some insight on how to use the tool for an endogamous population like the Ashkenazi Jews. I’m trying to use the tool for four different individuals who are 100% Ashkenazi and so far I’ve tried it on three of the four with the exact same results. I tried the default settings first, but got one large cluster with all the matches the system found. So then I tried every different combination of the number of matches and threshold levels I could think of and continued to get a single large cluster. I did not change any of the other defaults since they did not seem like they should or would affect the results, but obviously, if I’m wrong about that, please let me know. I’m not really sure what else to do or to try.
FYI, I also used the tool on someone who is about 50% Ashkenazi, and although I did get a large cluster among the results, there were a ton of clusters returned, including the specific one I was trying to find (which was not the large one), which may be related to this person’s Ashkenazi side of the family. So I’m not sure whether the issue I’m having is due to the endogamy or not, but thought it was worth reporting all my encounters with the tool.
When I heard about this tool, I was so hopeful, because it’s really hard to have any success with DNA research for Ashkenazi Jews between the large number of matches and the problem finding traditional research from Eastern Europe to confirm who the MRCA is (between language issues, WWII document destruction and document dispersal, the general lack of Jewish surnames prior to the early 1800s, etc).
I really hope you can help me figure out a way to make it work these individuals. Thanks!!
I have mild endogamy from groups who were in the same area for 800-1000 years. GA clustering and so on works reasonably well, but only because I know which of my ancestral lines might be entangled in such clusters, and therefore how to untangle them.
You are very brave trying clustering to split out different parts of a group with endogamy AND a severe bottleneck event in their past millenium.
Best of luck!
Hi Michelle,
EJ here from Genetic Affairs. You are correct that running the tool for an endogamous population is challenging.
First, the clustering does not work properly because only one (or a couple) cluster is identified. Next, the used probabilities are created based on non-endogamous relationships.
Perhaps if we clusters based on large triangulating segments, and have an updated probability table for endogamous populations, it would be possible to cluster (and perform subsequent AutoKinship) analyses. Up until then, you could look at other methods that use segments for clustering, like the AutoSegment tool (also available on GEDmatch!).
Thanks Roberta.
I’ve just been through the Genetic Affairs tools with my 23andMe results so I am looking forwards to trying it at GEDmatch. I think it’s a great tool for adoptees and for people starting out – the main problem being that someone starting out won’t know what it means.
At present I think newbies need to cut their teeth on Ancestry and MyHeritage first.
But I love it.
One question – what is the probability figure at AutoKinship about?
To me, one chance in 10^-18 is about the chance I have of winning the lottery twice in a row. Or a politician following through on every single promise. What does a bigger/smaller number mean? Why all of those extra digits when they are not in the technical term, “significant figures”?
GA clusters and AutoKinship won’t solve all of everyone’s problems all at once, but an “in” for one problem often works towards solving another.
People should try it.
Thanks Christopher,
Your question about the low probabilities is a valid one. I am not a statistician, but here is my take on things. As you know, only a handful of relationships have a probability of 100% giving a certain amount
of shared cM. As soon as you share less than a parent-child relationship, the probabilities are much lower than 100%.
To compare different scenarios, we need to consider every relationship (and therefore each probability linked to it) for a certain generated tree. A perfect tree consisting of 4 high probability (100% each) relationships would have hypothetical compound probability of 100% ==> 100% times 100% times 100% times 100%.
By having one relationship in there with a probability of 50%, you already have a compound probability of 50% ==> 100% times 100% times 100% times 50%.
And if these were only probabilities of 10%, it would be much lower ==> 10% times 10% times 10% times 10% = 0.01%
If your tree becomes very large, with some distant relationships, the compound probability becomes quite small. That’s why it’s only fair to analyze the generated tree by comparing it to the other generated trees and see how different they are. If one tree has a compound probability of 0.01% and another has 0.001%, the first one is ten times more likely as the second one (0.01/0.001).
I believe the same principle is being used by WATO, although the probabilities much be higher because they only look from the perspective of the test taker wheras AutoKinship employs relationships from every DNA match (including the test taker)
I’m trying to determine a set of 3rd great grandparents.
Probably too much to ask for, but these features would greatly be enhanced if they could be run on multiple kits simultaneously (eg., three 1st cousins), would they not?
Great idea.
very complicated_-way beyond my abilities!!!!!!!!!
I do not understand why my mother, daughter, aunt, and 1st cousins do not show up in any of my clusters. Nor do they show up in each others.
Close family members are not included in clusters because they would be in every cluster. Look at the thresholds.
hi susan, sorry for the late reply. If you increase the upper cM threshold limit, you should be able to see your close family members. However, this feature is perhaps best appreciated when trying to get to know an unknown match. By performing the analysis on the close DNA matches of that match, some hypotheses might be generated based on the outcome.
Hi,
In the description above regarding clusters, the options are described as a default and a dense, but then that the default is the dense. Which of the two is the recommended option for better matches?
Roberta, it is my understanding that Ancestry made some changes with their Pro Tools, creating a need for Genetic Affairs to update its software to work with the way Ancestry is set up now. Do you know if they have made the necessary updates so that it works with the new Ancestry version?
Please ask Genetic Affairs.
I get 5 auto trees, but they are blank. What does that mean?