Recently, I’ve received a number of questions about comparing people and haplogroups between 23andMe and Family Tree DNA. I can tell by the questions that a significant amount of confusion exists about the two, so I’d like to talk about both. In you need a review of “What is a Haplogroup?”, click here.
Haplogroup information and comparisons between Family Tree DNA information and that at 23andMe is not apples and apples. In essence, the haplogroups are not calculated in the same way, and the data at Family Tree DNA is much more extensive. Understanding the differences is key to comparing and understanding results. Unfortunately, I think a lot of misinterpretation is happening due to misunderstanding of the essential elements of what each company offers, and what it means.
There are two basic kinds of tests to establish haplogroups, and a third way to estimate.
Let’s talk about mitochondrial DNA first.
Mitochondrial DNA
You have a very large jar of jellybeans. This jar is your mitochondrial DNA.
In your jar, there are 16,569 mitochondrial DNA locations, or jellybeans, more or less. Sometimes the jelly bean counter slips up and adds an extra jellybean when filling the jar, called an insertion, and sometimes they omit one, called a deletion.
Your jellybeans come in 4 colors/flavors, coincidentally, the same colors as the 4 DNA nucleotides that make up our double helix segments. T for tangerine, A for apricot, C for chocolate and G for grape.
Each of the 16,569 jellybeans has its own location in the jar. So, in the position of address 1, an apricot jellybean is always found there. If the jellybean jar filler makes a mistake, and puts a grape jellybean there instead, that is called a mutation. Mistakes do happen – and so do mutations. In fact, we count on them. Without mutations, genetic genealogy would be impossible because we would all be exactly the same.
When you purchase a mitochondrial DNA test from Family Tree DNA, you have in the past been able to purchase one of three mitochondrial testing levels. Today, on the website, I see only the full sequence test for $199, which is a great value.
However, regardless of whether you purchase the full mitochondrial sequence test today, which tests all of your 16,569 locations, or the earlier HVR1 or HVR1+HVR2 tests, which tested a subset of about 10% of those locations called the HyperVariable Region, Family Tree DNA looks at each individual location and sees what kind of a jellybean is lodged there. In position 1, if they find the normal apricot jellybean, they move on to position 2. If they find any other kind of jellybean in position 1, other than apricot, which is supposed to be there, they record it as a mutation and record whether the mutation is a T,C or G. So, Family Tree DNA reads every one of your mitochondrial DNA addresses individually.
Because they do read them individually, they can also discover insertions, where extra DNA is inserted, deletions, where some DNA dropped out of line, and an unusual conditions called a heteroplasmy which is a mutation in process where you carry some of two kinds of jellybean in that location – kind of a half and half 2 flavor jellybean. We’ll talk about heteroplasmic mutations another time.
So, at Family Tree DNA, the results you see are actually what you carry at each of your individual 16,569 mitochondrial addresses. Your results, an example shown below, are the mutations that were found. “Normal” is not shown. The letter following the location number, 16069T, for example, is the mutation found in that location. In this case, normal is C. In the RSRS model of showing mitochondrial DNA mutations, this location/mutation combination would be written as C16069T so that you can immediately see what is normal and then the mutated state. You can click on the images to enlarge.
Family Tree DNA gives you the option to see your results either in the traditional CRS (Cambridge Reference Sequence) model, above, or the more current Reconstructed Sapiens Reference Sequence (RSRS) model. I am showing the CRS version because that is the version utilized by 23andMe and I want to compare apples and apples. You can read about the difference between the two versions here.
Defining Haplogroups
Haplogroups are defined by specific mutations at certain addresses.
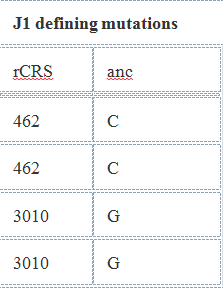
For example, the following mutations, cumulatively, define haplogroup J1c2f. Each branch is defined by its own mutation(s).
| Haplogroup | Required Mutations |
| J | C295T, T489C, A10398G!, A12612G, G13708A, C16069T |
| J1 | C462T, G3010A |
| J1c | G185A, G228A, T14798C |
| J1c2 | A188G |
| J1c2f | G9055A |
You can see, below, that these results, shown above, do carry these mutations, which is how this individual was assigned to haplogroup J1c2f. You can read about how haplogroups are defined here.
At 23andMe, they use chip based technology that scans only specifically programmed locations for specific values. So, they would look at only the locations that would be haplogroup producing, and only those locations. Better yet if there is one location that is utilized in haplogroup J1c2f that is predictive of ONLY J1c2f, they would select and use that location.
This same individual at 23andMe is classified as haplogroup J1c2, not J1c2f. This could be a function of two things. First, the probes might not cover that final location, 9055, and second, 23andMe may not be utilizing the same version of the mitochondrial haplotree as Family Tree DNA.
By clicking on the 23andMe option for “Ancestry Tools,” then “Haplogroup Tree Mutation Mapper,” you can see which mutations were tested with the probes to determine a haplogroup assignment. 23andMe information for this haplogroup is shown below. This is not personal information, meaning it is not specific to you, except that you know you have mutations at these locations based on the fact that they have assigned you to the specific haplogroup defined by these mutations. What 23andMe is showing in their chart is the ancestral value, which is the value you DON’T have. So your jelly bean is not chocolate at location 295, it’s tangerine, apricot or grape.
Notice that 23andMe does not test for J1c2f. In addition, 23andMe cannot pick up on insertions, deletions or heteroplasmies. Normally, since they aren’t reading each one of your locations and providing you with that report, missing insertions and deletions doesn’t affect anything, BUT, if a deletion or insertion is haplogroup defining, they will miss this call. Haplogroup K comes to mind.
23andMe never looks at any locations in the jelly bean jar other than the ones to assign a haplogroup, in this case,17 locations. Family Tree DNA reads every jelly bean in the jelly bean jar, all 16,569. Different technology, different results. You also receive your haplogroup at 23andMe as part of a $99 package, but of course the individual reading of your mitochondrial DNA at Family Tree DNA is more accurate. Which is best for you depends on your personal testing goals, so long as you accurately understand the differences and therefore how to interpret results. A haplogroup match does not mean you’re a genealogy match. More than one person has told me that they are haplogroup J1c, for example, at Family Tree DNA and they match someone at 23andMe on the same haplogroup, so they KNOW they have a common ancestor in the past few generations. That’s an incorrect interpretation. Let’s take a look at why.
Matches Between the Two
23andMe provides the tester with a list of the people who match them at the haplogroup level. Most people don’t actually find this information, because it is buried on the “My Results,” then “Maternal Line” page, then scrolling down until your haplogroup is displayed on the right hand side with a box around it.
Those who do find this are confused because they interpret this to mean they are a match, as in a genealogical match, like at Family Tree DNA, or like when you match someone at either company autosomally. This is NOT the case.
For example, other than known family members, this individual matches two other people classified as haplogroup J1c2. How close of a match is this really? How long ago do they share a common ancestor?
Taking a look at Doron Behar’s paper, “A “Copernican” Reassessment of the Human Mitochondrial DNA Tree from its Root,” in the supplemental material we find that haplogroup J1c2 was born about 9762 years ago with a variance of plus or minus about 2010 years, so sometime between 7,752 and 11,772 years ago. This means that these people are related sometime in the past, roughly, 10,000 years – maybe as little as 7000 years ago. This is absolutely NOT the same as matching your individual 16,569 markers at Family Tree DNA. Haplogroup matching only means you share a common ancestor many thousands of years ago.
For people who match each other on their individual mitochondrial DNA location markers, their haplotype, Family Tree DNA provides the following information in their FAQ:
-
- Matching on HVR1 means that you have a 50% chance of sharing a common maternal ancestor within the last fifty-two generations. That is about 1,300 years.
- Matching on HVR1 and HVR2 means that you have a 50% chance of sharing a common maternal ancestor within the last twenty-eight generations. That is about 700 years.
- Matching exactly on the Mitochondrial DNA Full Sequence test brings your matches into more recent times. It means that you have a 50% chance of sharing a common maternal ancestor within the last 5 generations. That is about 125 years.
I actually think these numbers are a bit generous, especially on the full sequence. We all know that obtaining mitochondrial DNA matches that we can trace are more difficult than with the Y chromosome matches. Of course, the surname changing in mitochondrial lines every generation doesn’t help one bit and often causes us to “lose” maternal lines before we “lose” paternal lines.
Autosomal and Haplogroups, Together
As long as we’re mythbusting here – I want to make one other point. I have heard people say, more than once, that an autosomal match isn’t valid “because the haplogroups don’t match.” Of course, this tells me immediately that someone doesn’t understand either autosomal matching, which covers all of your ancestral lines, or haplogroups, which cover ONLY either your matrilineal, meaning mitochondrial, or patrilineal, meaning Y DNA, line. Now, if you match autosomally AND share a common haplogroup as well, at 23andMe, that might be a hint of where to look for a common ancestor. But it’s only a hint.
At Family Tree DNA, it’s more than a hint. You can tell for sure by selecting the “Advanced Matching” option under Y-DNA, mtDNA or Family Finder and selecting the options for both Family Finder (autosomal) and the other type of DNA you are inquiring about. The results of this query tell you if your markers for both of these tests (or whatever tests are selected) match with any individuals on your match list.
Hint – for mitochondrial DNA, I never select “full sequence” or “all mtDNA” because I don’t want to miss someone who has only tested at the HVR1 level and also matches me autosomally. I tend to try several combinations to make sure I cover every possibility, especially given that you may match someone at the full sequence level, which allows for mutations, that you don’t match at the HVR1 level. Same situation for Y DNA as well. Also note that you need to answer “yes” to “Show only people I match on all selected tests.”
Y-DNA at 23andMe
Y-DNA works pretty much the same at 23andMe as mitochondrial meaning they probe certain haplogroup-defining locations. They do utilize a different Y tree than Family Tree DNA, so the haplogroup names may be somewhat different, but will still be in the same base haplogroup. Like mitochondrial DNA, by utilizing the haplogroup mapper, you can see which probes are utilized to determine the haplogroup. The normal SNP name is given directly after the rs number. The rs number is the address of the DNA on the chromosome. Y mutations are a bit different than the display for mitochondrial DNA. While mitochondrial DNA at 23andMe shows you only the normal value, for Y DNA, they show you both the normal, or ancestral, value and the derived, or current, value as well. So at SNP P44, grape is normal and you have apricot if you’ve been assigned to haplogroup C3.
As we are all aware, many new haplogroups have been defined in the past several months, and continue to be discovered via the results of the Big Y and Full Y test results which are being returned on a daily basis. Because 23andMe does not have the ability to change their probes without burning an entirely new chip, updates will not happen often. In fact, their new V4 chip just introduced in December actually reduced the number of probes from 967,000 to 602,000, although CeCe Moore reported that the number of mtDNA and Y probes increased.
By way of comparison, the ISOGG tree is shown below. Very recently C3 was renamed to C2, which isn’t really the point here. You can see just how many haplogroups really exist below C3/C2 defined by SNP M217. And if you think this is a lot, you should see haplogroup R – it goes on for days and days!
How long ago do you share a common ancestor with that other person at 23andMe who is also assigned to haplogroup C3? Well, we don’t have a handy dandy reference chart for Y DNA like we do for mitochondrial – partly because it’s a constantly moving target, but haplogroup C3 is about 12,000 years old, plus or minus about 5,000 years, and is found on both sides of the Bering Strait. It is found in indigenous Native American populations along with Siberians and in some frequency, throughout all of Asia and in low frequencies, into Europe.
How do you find out more about your haplogroup, or if you really do match that other person who is C3? Test at Family Tree DNA. 23andMe is not in the business of testing individual markers. Their business focus is autosomal DNA and it’s various applications, medical and genealogical, and that’s it.
Y-DNA at Family Tree DNA
At Family Tree DNA, you can test STR markers at 12, 25, 37, 67 and 111 marker levels. Most people, today, begin with either 37 or 67 markers.
Of course, you receive your results in several ways at Family Tree DNA, Haplogroup Origins, Ancestral Origins, Matches Maps and Migration Maps, but what most people are most interested in are the individual matches to other people. These STR markers are great for genealogical matching. You can read about the difference between STR and SNP markers here.
When you take the Y test, Family Tree DNA also provides you with an estimated haplogroup. That estimate has proven to be very accurate over the years. They only estimate your haplogroup if you have a proven match to someone who has been SNP tested. Of course it’s not a deep haplogroup – in haplogroup R1b it will be something like R1b1a2. So, while it’s not deep, it’s free and it’s accurate. If they can’t predict your haplogroup using that criteria, they will test you for free. It’s called their SNP assurance program and it has been in place for many years. This is normally only necessary for unusual DNA, but, as a project administrator, I still see backbone tests being performed from time to time.
If you want to purchase SNP tests, in various formats, you can confirm your haplogroup and order deeper testing.
You can order individual SNP markers for about $39 each and do selective testing. On the screen below you can see the SNPs available to purchase for haplogroup C3 a la carte.
You can order the Geno 2.0 test for $199 and obtain a large number of SNPs tested, over 12,000, for the all-inclusive price. New SNPs discovered since the release of their chip in July of 2012 won’t be included either, but you can then order those a la carte if you wish.
Or you can go all out and order the new Big Y for $695 where all of your Y jellybeans, all 13.5 million of them in your Y DNA jar are individually looked at and evaluated. People who choose this new test are compared against a data base of more than 36,000 known SNPs and each person receives a list of “novel variants” which means individual SNPs never before discovered and not documented in the SNP data base of 36,000.
Don’t know which path to take? I would suggest that you talk to the haplogroup project administrator for the haplogroup you fall into. Need to know how to determine which project to join, and how to join? Click here. Haplogroup project administrators are generally very knowledgeable and helpful. Many of them are spearheading research into their haplogroup of interest and their knowledge of that haplogroup exceeds that of anyone else. Of course you can also contact Family Tree DNA and ask for assistance, you can purchase a Quick Consult from me, and you can read this article about comparing your options.
______________________________________________________________
Disclosure
I receive a small contribution when you click on some of the links to vendors in my articles. This does NOT increase the price you pay but helps me to keep the lights on and this informational blog free for everyone. Please click on the links in the articles or to the vendors below if you are purchasing products or DNA testing.
Thank you so much.
DNA Purchases and Free Transfers
- Family Tree DNA
- MyHeritage DNA only
- MyHeritage DNA plus Health
- MyHeritage FREE DNA file upload
- AncestryDNA
- 23andMe Ancestry
- 23andMe Ancestry Plus Health
- LivingDNA
Genealogy Services
Genealogy Research
- Legacy Tree Genealogists for genealogy research
Discover more from DNAeXplained - Genetic Genealogy
Subscribe to get the latest posts sent to your email.











Awesome Roberta! Thank you.
Carla Wallain Davis
________________________________
I love the jellybean analogy.
I have one case where matching (or NOT matching) haplogroup can be done within autosomal DNA. On my father line, our surname project has identified 4 sons of a MRCA from the early 1700s. We don’t know his parents. Our haplogroup is I-M223. When Ancestry shows a distant cousin match to another tested line whose haplogroup is R1b, I know the “match” is almost certainly IBS.
Thanks for reminding me about advanced testing page;
I can understand having no common FMS, HVR1, HVR2 and FF matches as we are talking several hundreds of years or more than 1000 years.
I have several Y STR matches including 66/67 (different surnames) matches and some have done the FF test with “underwhelming” results; The TIP test says the 66/67 match is 90% likely to have had a common ancestor within 8 generations past. But I have no matches with the FF results……
Would we not expect to have some autosomal (FF) matches in in there at least at the 5th to remote level???
Steve in Oro Valley
Hi Steve, both the TIP and the cousin predictor use statistics based on averages. http://dna-explained.com/2012/11/29/averages-tip-calculator-and-one-size-fits-all/ and http://dna-explained.com/2014/02/19/generational-inheritance/ and http://dna-explained.com/2013/10/21/why-are-my-predicted-cousin-relationships-wrong/
I know how frustrating this is.
For those who are searching for their MRCA through Y-DNA, I strongly recommend FTDNA’s Big Y, and wouldn’t even consider 23andMe. I’ve tested with both as much as I can and found my terminal SNP through FTDNA’s Deep Clade test a couple of years ago. To show you the difference, FTDNA reported that my terminal SNP was L176.2, but that I also tested positive for L147.3, which is still under research according to ISOGG. I share this latter SNP with just one other person who estimated that we share a MRCA over 500 years ago. And that’s way down on the tree! I hope Big Y comes up with closer resolution. My haplogroup at L176.2 is reported by ISOGG as R1b1a2a1a2a1b. Whew!
23andMe had reported my haplogroup as R1b1b2a1a, which is not even listed under the ISOGG tree, and according to 23andMe’s Mapper, the “terminal” SNPs they tested were 4 levels upstream! They missed P312, >DF27, >Z195; no hope for finding any matches there. 23andMe clearly focused on medical testing (until the FDA shut that off) and not genetic genealogy.
Roberta, I saw at FTDNA that they can ‘retest?’ your 23&me – Autosomal Transfers for $69, should I do this OR should I do the whole thing over at FTDNA for the $199? Decisions Decisions….
The 23andMe is a transfer of your raw data file, for $69. You download it from 23andMe and upload it to FTDNA. If you decide to have FTDNA retest your DNA, its $99, not $199. If you tested at 23andMe in December 2013 or after, on the V4 chip, you have to retest. If you tested on the V2 chip so very early at 23andMe, you have to retest too.
I did mine in late Oct/early Nov 2013. So, I just upload my full sequence from 23&me, $69, and they do a full reading of it? I know, questions, send me a bill & I’ll send you a check, as this is turning into a consult, isn’t it…..I just didn’t understand if I should retest or if since I already did the 23&me, what would be the best plan for me to get the best results.
It’s just that this article has me thinking, as I did the 23&me because of the V2 chip thing, I got my full genome back on the 20th of Nov 2013. I had my Sorenson test manually transferred on Nov 11th, that is when I call FT after I found out that they had bought all the Sorenson data and it took them a bit to find me, as I had lost that print out in storage [one of the boxes of paper that the vermin decided to snack on when they got tired of my clothes & furniture]. From the Sorenson transfer, I am H, and 23&me, I am J1c1b1a. I didn’t realize until reading the above that 23&me wasn’t that thorough. My brother, Richard W Naylor Sr., did the FTDNA for his Y and then I sent him a 23&me kit. Send me your mailing address in a private email, and I’ll shoot you a check.
I have been diligently adding the new info from your 52/52 into my tree for the grandkids. I am going to print them out a book of the TN/VA ancestors. I printed out the “Crazy Aunts” for JD to take offshore, and I am going to include to for Amos the kids, I got such a chuckle from it. Minnie was a trip! I miss the old gal.
Great Article Thanks. Have you or do you plan to write a similar article explaining what if any differences exist between kits from FTDNA and AncestryDNA (part of Ancestry.com where I have my family tree posted)? I have done (in 2010) 12-Marker YDNA at FTDNA + awaiting Autosomal results from AncestryDNA. I just ordered a Autosomal kit for my wife – also from AncestryDNA. I have principally chosen these respective companies based on ability to post results at both companies websites and see what matches are listed. If I had done Autosomal test at FTDNA, I was advised by AncestryDNA that I can not upload those result to AncestryDNA site. I am an only child – parents are dead and have no contact with any family members. I know I have cousins, etc but have no idea where they are or if they are alive. Principal goal is to identify a person(s) that is a relative (either maternal or paternal) and make contact with that person or relative of that person to hopefully learn more about my family. Any guidance would be appreciated regarding DNA testing and best company for each.
Hi Harry. I did a comparison between Ancestry.com and Family Tree DNA some time back. It’s much less relevant now because Ancestry is no longer selling their Y DNA and mito tests, just their autosomal. Here’s the link to the Y article: http://dna-explained.com/2012/08/14/y-dna-family-tree-dna-vs-ancestry/ Here’s the article about mtdna: http://dna-explained.com/2012/07/18/the-trouble-with-ancestry-com-matches/
Regarding the autosomal tests at both companies, Ancestry give you no tools like the Chromosome browser, the matrix, no admin tools like the group matching tools, nothing…..so other than they match you with other trees, as far as I’m concerned, they have nothing to offer in terms of actually proving your connection. It’s a “trust me” or an assumed type of situation, which as far as I’m concerned, it entirely unacceptable. So, IMHO, there is little to compare. This article was written about what was needed, and why, over a year ago and Ancestry has not responded in any positive fashion. http://dna-explained.com/2013/03/24/ancestry-needs-another-push-chromosome-browser/
Thank you very much for articles and prompt response. I am continuing to learn about genealogy in general and how DNA testing can work with it. Thanks again
Harry
Another advantage to FTDNA (besides the many tools on their website) is being able to use the DNAgedcom.com, the wonderful tool you wrote about on Jan 9 2014 and which I immediately loaded with my atDNA results. A wonderful tool, and thank you for letting us know about it.
My brother is one of those who they were unable to predict a haplogroup for and are doing the free SNP test at FTDNA – I gave him a 67 marker test and atDNA for Xmas. Thank you for helping me understand the why of this in today’s post. My father was a member of the Odd Fellows. Guess he really was!!
This article came at a very crucial time in my DNA research. I recently had a supposedly second paternal cousin C tested 23 andme. He matched all paternal cousins less than expected. and had a Haplogroup of
Hi, I see your experience sounds similar to ours.
My first paternal cousin tested at FTDNA and was assigned Haplogroup T. A free backbone test was done and a m70 was assigned. 23andme assigned him a T.
Another paternal cousin, C who 23 andme assigned Haplogroup R1b1b2a1a2f. When I contacted the surname project, he questioned the accuracy on the 23and me but I do not. I have tested this same first cousin and a fifth half paternal cousin on SMFG and the only variance as far as I can determine is on marker DYS 456 by one number. The first paternal cousin is 13 . The fifth half paternal cousin is 14. The common paternal ancestor for all these males should be my great paternal grandfather.
I have several kits on 23 andme from this family and Paternal cousin, C who 23 andme assigned Haplogroup R1b1b2a1a2f matches all his paternal cousins less than he should.
This had been further complicated by 23andme changing to chip v and I am unable to upload Cousin C autosome result to FTDNA to compare to know matches coming though the paternal great grandmother. These are all back by a good solid paper trail. Very confused
The only chip compatible with FTDNA is version 3. So no version 4 files since December 2013 will work.
Roberta,
I want to let you know that your blog post is listed in today’s Fab Finds post at http://janasgenealogyandfamilyhistory.blogspot.com/2014/03/follow-friday-fab-finds-for-march-28.html
Have a wonderful weekend!
Thank you Jana.
Pingback: Haplogroup Projects | DNAeXplained – Genetic Genealogy
I why do all transfers from services to services and all services for testing, assume we ( the tested ) have every basal marker for the haplogroup we belong to even though we do not?
Pingback: Autosomal DNA 2015 – Which Test is the Best? | DNAeXplained – Genetic Genealogy
Hi Roberta. I did full sequence at FTDNA and just recently did the 23 test and received the exact halogroup as my full sequence. I just need to know if that will be the case if I do testing for someone else because I don’t want to spend that much to get my paternal grandfather’s mothers halogroup. (Or anyone else for that matter on other lines) Thoughts?
That’s not always the case based on the DNA and the probe locations tested for the haplogroup. What you don’t get from 23andMe are all of the mutations and matches.
Pingback: DNAeXplain Archives – Introductory DNA | DNAeXplained – Genetic Genealogy
Hello, i tested my y-25 and it shows that my haplogroup is C-M130 (too general) and i have a question what if i order y-111 will it show more accurate SNP result?
Thanks
No, upgrading your STR markers won’t clarify your haplogroup. You need to order SNPs to do that. However, there are a variety of ways to do that. If you are a member of a haplogroup project, the admin would probably be glad to help you figure out what is best for your situation. For the most advanced test, the Big Y, you must upgrade to 37 markers first. These two tests work together to provide deep ancestry for you.
During my research of which FTDNA Kit to order I was disappointed to find a significant cost between the 37 Marker, the 67 Marker, and the 111 Marker, so I’m briefly going to tell you what I want and hope you will advise me which Kit I should order… I specifically want to discover the haplogroup and what part of the world that my ancient ancestors lived thousands of years ago. So which Kit should I order and please understand that I already know much of my ancestry from five and six generations back, so that’s not the most important thing to me in choosing a kit.
Unfortunately, everyone wants that and it’s not quite as easy as that. You get a haplogroup estimate with any STR marker test purchase. If that’s good enough for what you want, then any purchase should suffice. However, with each higher level, your match mapping gets more granular, hopefully, so you can see where the people you match the most closely originated, as best they know. If you want more information about your deeper ancestry, you can purchase SNP panels after your STR markers are done and your haplogroup has been predicted. You cant purchase those without the haplogroup prediction. If you want the whole enchilada, then take the Big Y test which gives you all of your SNPs and novel variants, but because it’s a research type of test, new SNPs may be discovered that we don’t yet know where go on the tree. So, I guess I would say that the process of leaning where your ancestors were from is not a place but a path and all of the information from your Y chromosome is needed to understand that path. It’s fluid and we’re still learning about the finer pieces.
I also am curious about the older relationships. I know at least to my grandfather, and his father. FT offers the Y-DNA12 , which, reportedly will show likely related to 29 generation. More expensive tests show lower numbers of generations. Whats up with that?
You’re misunderstanding. The 12 marker can’t show close generations because there aren’t enough markets. The more markers, the more refined the test and the closer in time the test can tell you. That’s what you want in a y DNA test.
Close generations I already know. So the 12 marker test wouldn’t tell me anything I don’t already know. Why does it claim to be able to do 29 generations? Just a cursory overview then? Would it give me 29 times 20 years (580 years) worth of background? We know nothing previous to my great grandfather anyway.
Ok, let’s try this another way. At 12 markers you may match 1000 men with all different surnames including a few that are similar to yours. That’s cause those people are related to you but very far back in time. Want to know which of those people are related in a genealogically relevant timeframe? Order more markers. Even the same surname may be from different family lines hundreds to thousands of years ago. Families like Miller and smith know that all too well. Bottom line. If you don’t test you’ll never know what you don’t know. Today we really don’t recommend less that 37 markers.
Mrs Roberta Estes, I already got my Big Y DNA Results and they told me when i belongs to Haplogroup O-CTS11856 / O-M117 +M133 or O-CTS5492 from Geno 2,0. Frankly i don’t really understand with my Y Hg O-M133 because this Y Hg seems the “Youngest” Y Hg O’s or what? I have no idea…….
I would be glad to do a quick consult with you. http://www.dnaxplain.com/shop/features.aspx
I did YDNA-67 and matched extremely high to a few Fergusons (like myself) and other names (Lawson, Alexander, Maier, Holland). Ranging from 64-67/67. Seemed to have ruled out external paternal events…project manager for Clan Ferguson believes it’s based upon older generations changing their last name….and that my YDNA just never really mutated much…if it did mutate, it may have mutated back and forth to seem as if their isn’t much change….???
for Ancestry and me I tried to process my YDNA twice and they rejected my sample saying it wasn’t sufficient…despite, AncestryDNA and FTDNA never having a problem. Apparently Ancestry and me as you kind of explained make their determinations based on different criteria. My haplogroup is U152 terminal.
Interested to hear what anyone has to say on this.
Hi Roberta, hopefully you still have a little patience for us newbies figuring out the complex world of dna (at least to those starting from scratch)! My mother and father are fortunately still both living. My interest at this time is to get the most accurate and affordable results.
From what I’m gathering from your article, it’s a 50% chance of accuracy whether I use my own dna and get the mitochondrial as it would be to test my mother and father seperately. Am I right? In my brain, testing my father’s Y and moms mitochondrial might give me more accuracy, but I may be way off.
I know my lineage on dad’s side up to about 300 yrs and moms too, but not much more then grandparents and not for instance who my second and third cousins living may be, so these results would be interesting to learn.
I want to get my parents this gift for Christmas but am wondering if I should just do myself and get enough info to share with them since they are from 2 different parts of Europe for certain, so defining which parts is my second desire, but that also would make it hard to figure out which come from maternal and paternal.
Thanks in advance for any help you are willing to offer!
I’m not sure what you mean about the 50% accuracy. The Y DNA and the mitochondrial DNA are passed intact between generations, except for any mutations that might happen. It’s autosomal DNA where you only receive half of your mother’s DNA and half of your father’s. However, your father inherited his mother’s mitochondrial DNA, which you did not. So if you test your father, you can get his Y line, which females don’t carry, his mitochondrial, which his children don’t carry, and his autosomal which will help you determine which “side” of your family your own autosomal matches are from. If you test your mother, you and she carry the same mitochondrial DNA, but her test will help sort your autosomal matches into “sides” of the family that they came from. So, my recommendation would be Y, mtDNA and Family Finder (autosomal) for your Dad. Mitochondrial and Family Finder for your mother and Family Finder for you. There are coupons available in this article in addition to the sale prices. https://dna-explained.com/2016/11/22/family-tree-dna-2016-holiday-sale/
Pingback: New Native American Mitochondrial DNA Haplogroups | DNAeXplained – Genetic Genealogy
Pingback: Mitochondrial DNA Build 17 Update at Family Tree DNA | DNAeXplained – Genetic Genealogy
Pingback: Concepts – Percentage of Ancestors’ DNA | DNAeXplained – Genetic Genealogy
Pingback: Which DNA Test is Best? | DNAeXplained – Genetic Genealogy
Pingback: LivingDNA Product Review | DNAeXplained – Genetic Genealogy
Thank you for this DNA insight!
Would 2 brothers get the same information from their Y-dna and Mitochondrial Dna tests? Would they have the same mutations you referred to above? I’m trying to decide if I need to test both brothers Y-DNA and Mitochondrial, but if it would be identical I shouldn’t need both of them to take these tests. Am I correct thinking this?
No, you don’t need both brothers to test Y and mitochondrial DNA, but you DO need them both to test autosomal DNA, unless your parents are both living.
Hi Roberta,
This question has more to do with what I should expect to learn from my 23andme DNA test. I plan on trying other tests as well, but received this one as a Christmas Gift. My mother told me who my biological father was when I was 12 (1992-93). His now wife wanted a DNA test done but changed their location last minute…long story short DNA test came back saying he was not my father. Aside from the possibility of tampering, I have read where sometimes mothers and son’s samples get mixed up (they don’t test for X,Y) and/or also only testing at 2 locations (not accounting for possible mutations). Will this test help me find close relatives, and if so, will it distinguish between maternal and paternal relatives? I am a 37 year old male. Thank you for your help.
~Dustin
Hi Dustin. There are several questions that need to be asked. I would be more than happy to do a personal consult for you. http://www.dnaxplain.com/shop/features.aspx
A third cousin did a yDNA test on Ancestry back when they were still doing that (because his sister was an Ancestry member already so just did it there rather than really checking out FT). His results were then “converted” to the FT standard and put on ySearch where we found matches who had tested on FT. Are matches found this way reliable? (The surname is right, and the connections make sense, though we haven’t found the paper trail yet.) I’ve been using one of the perfect matches (on 34 mutual markers tested) as a surrogate for finding new FT matches, and want to confirm that this is valid.
Thanks.
Marian
Yes, it’s valid. However, everyone doesn’t upload to YSearch so if he can retest at FTDNA, that would probably produce more matches.
Thanks. I’m working on that, but meanwhile just wanted confirmation that I am on the right track. Now that only FT is doing the yDNA, I think fewer people are bothering with ySearch. but fortunately the ySearch match who tested on FT is keeping me in the loop with what is going on there.
I guess I am concerned that my FT Family Finder matches don’t include any of the same people as the ySearch match’s Family Finder or yDNA matches. (Ancestry and 23andMe both say that I match with my cousin.)
Since more people seem to be testing with Ancestry and 23andMe (good advertising, I think!), I have been working on trying to filter matches there to find potential yDNA matches.
Y Search doesn’t have autosomal, so it won’t show that. You need to compare Family Finder with Family Finder at Family Tree DNA. Ancestry doesn’t have Y. So this is apples and oranges.
Sorry. I was unclear. I am taking the person who is my cousin’s yDNA match on ySearch and comparing his FTDNA Family Finder matches with my FTDNA Family Finder matches. (We don’t show up as matching each other.) Actually, now that I am comparing all 5000+ of his matches with all 3000+ of mine (thank you, Excel! It would have been so much easier if the csv download had included ID#s!!) I am finding some mutual matches.
Best explanation ever. I finally understand some things I didn’t before.
Hi Ms. Estes,
I’m hoping you could answer my question or guide me as to what I should do next. In 2015 I had my male first cousin do the FTDNA Y-DNA37 test as I was interested in tracing my maternal grandfather’s line. FTDNA predicted the Y Haplogroup to be IM253. Which surprised me because as far as we knew my grandparents were from Japan. Fast forward to June 2018, some of my friends suggested I do DNA testing through 23 and me which I did and I decided that I would also ask the same male cousin to do this test as well. When he got the results back it stated his Y Haplogroup was O-F8. So I am confused. Which Y Haplogroup is correct?
Does he match you autosomally the way you would expect? Did you submit any other tests at the same time that could have been switched? I can’t answer the question, but I would suggest digging further. On the Y test, dies he match men with I or O haplogroups?
Hi Ms. Estes, for some reason I never got to see your answer until now. I am still confused. In answer to your first question, yes. No to the second question. To the third question, how do I tell who he matches with?
I had another male cousin on my maternal grandfather’s side (whose father was the son of my maternal grandfather’s brother) do the DNA test on 23 and Me and he showed O-F8 as his Y haplogroup. Should I assume that is the correct Y haplogroup, or should I also have him do a DNA test through FTDNA?
Pingback: Family Tree DNA’s Mitochondrial Haplotree | DNAeXplained – Genetic Genealogy
Hi. About 2 years ago I had 4 male relatives on 23andme with my last name. One was J-M267 and after 23andme updated the Y haplogroups around 2 years ago he remained as a J-M267. Another went from J-M267 to J1a (CTS5368/Z2215) after the update. The other two on 23andme that had J1a and J-P58 both remained as J1a and J-P58 after the update. I have one relative with my same surname on FTDNA that has J-PF4799 (L-862/Z2331) and I just found out he tested with Genographic 2.0 and uploaded his test to FTDNA.
My question is If 23andme is able to test enough SNPs to give someone a J-P58 even before the update 2 years ago why didnt the other 3 males went from J1 or J1a to J-P58 when the update took place?. Was it an issue of not enough data or chip? Or could we simply have different lineages here?
J1a is a mitochondrial haplogroup. Also, different vendors use different chips. 23andMe uses a customized medical chip.
Hi Roberta. Thanks for your prompt reply. J1a is a Y haplogroup more commonly known today as J-CTS5368/Z2215. I believe in the past it was called J1e but in the most recent Haplogroup J Tree its now called J1a as you can see here https://docs.google.com/spreadsheets/d/1CODtnxuvXZp1uxbJY53KUy0uT3ranaqE7N-nuqpAX-E/edit?fbclid=IwAR1KwxFtTyToazDp0qMDqsfDpqadNtkWWfLGYB_Y7hkDSs9XmWgDCCBi1BE#gid=1603190133.
I understand the different vendors use different chips but the 4 male testers I am referring to all tested in 23andme.
Could it be that the male tester that got J-P58 which is a subclade 3-5 levels down from J1-J1a just happen to be lucky that 23andme was able to extract a lot more dna data from him whereas those that remained as J1 and J1a after the update 2 years ago just werent as lucky? Or are we talking about two different lineages here that happen to share the same surname?
Right, but the same vendor isn’t going to use both the old and new style haplogroup at the same time.
Correct. 23and now uses J-M267 previously J1 and J-CTS5368 previously J1a. J-P58 used to be J1e I believe.
Is it possible that those 3 testers that remained as J-M267 and J-CTS5368 could still be from the same lineage as the tester who got J-P58? J-P58 is about 3-5 levels down from J-CTS5368…
Part of that depends on what chip they tested on. If you test at Family Tree DNA, they have matching and high resolution haplogroups available.
Pingback: Haplogroup Matching: What It Does (and Doesn’t) Mean | DNAeXplained – Genetic Genealogy
I think that 23andme is a great value, you get a lot of bang for your buck, but at least with haplogroup matching, I wasn’t satisfied. I got a different result (R-L1335 or Scots Modal) from my father, brother, and son (CTS241 or DF13). I then did the Big Y 700 with FTDNA and I got BY69501, which is a branch of R-Z3006, or Clan Colla. They are both downstream of DF13, but on completely different branches. I went back and forth with 23andme about their error, but they would respond with copied and pasted boilerplate messages about L1335 being downstream so there was no inconsistency. I guess quality control isn’t a priority for them.
23andme has my paternal haplogroup at I-S6687, but Family Tree has my paternal haplogroup at I-M170 for Y37. So, I-S6687 is only predicted due to the defining mutations as you mentioned, whereas I-M170 though not as ‘deep’ is more accurate due to the processes used. Because of 23andMe process, could the I-S6687 be inaccurate due to more than one subgroup having similar mutations at the same location? My reason for asking is that I am not willing to pay for additional levels or SNP testing if it could bare the same results.
If you want a more refined haplogroup, do the Big Y. It’s definitive and will put you as far down the tree as you can go.