After my introductory article, Y DNA: Part 1 – Overview, I received several questions about terminology, so this second article will be a dictionary or maybe more like a wiki. Many terms about Y DNA apply to mitochondrial and autosomal as well.
Haplogroup – think of your Y or mitochondrial DNA haplogroup as your genetic clan. Haplogroups are assigned based on SNPs, specific nucleotide mutations that change very occasionally. We don’t know exactly how often, but the general schools of thought are that a new SNP mutation on the Y chromosome occurs someplace between every 80 and 145 years. Of course, those would only be averages. I’ve as many as two mutations in a father son pair, and no mutations for many generations.

Y DNA haplogroups are quite reliably predicted by STR results at Family Tree DNA, meaning the results of a 12, 25, 37, 67 or 111 marker tests. Haplogroups are only confirmed or expanded from the estimate by SNP testing of the Y chromosome. Predictions are almost always accurate, but only apply to the upper level base haplogroups. I wrote about that in the article, Haplogroups and the Three Brothers.
Haplogroups are also estimated by some companies, specifically 23andMe and LivingDNA who provide autosomal testing. These companies estimate Y and mitochondrial haplogroups by targeting certain haplogroup defining locations in your DNA, both Y and mitochondrial. That doesn’t mean they are actually obtaining Y and mtDNA information from autosomal DNA, just that the chip they are using for DNA processing targets a few Y and mitochondrial locations to be read.
Again, the only way to confirm or expand that haplogroup is to test either your Y or mitochondrial DNA directly. I wrote about that in the article Haplogroup Comparisons Between Family Tree DNA and 23andMe and Why Different Haplogroup Results?.
Nucleotide – DNA is comprised of 4 base nucleotides, abbreviated as T (Thymine), A (Adenine), C (Cytosine) and G (Guanine.) Every DNA address holds one nucleotide.
In the DNA double helix, generally, A pairs with T and C pairs with G.

Looking at this double helix twist, green and purple “ladder rungs” represent the 4 nucleotides. Purple and green and have been assigned to one bonding pair, either A/T or C/G, and red and blue have been assigned to the other pair.
When mutations occur, most often A or T are replaced with their paired nucleotide, as are C and G. In this example, A would be replaced with T and vice versa. C with G and vice versa.
Sometimes that’s not the case and a mutation occurs that pairs A with C or G, for example.
For Y DNA SNPs, we care THAT the mutation occurred, and the identity of the replacing nucleotide so we know if two men match on that SNP. These mutations are what make DNA in general, and Y DNA in particular useful for genealogy.
The rest of this nucleotide information is not something you really need to know, unless of course you’re playing in the jeopardy championship. (Yes, seriously.) The testing lab worries about these things, as well as matching/not matching, so you don’t need to.
SNP – Single nucleotide polymorphism, pronounced “snip.” A mutation that occurs when the nucleotide typically found at a particular location (the ancestral value) is replaced with one of the other three nucleotides (the derived value.) SNPs that mutate are called variants.
In Y DNA, after discovery and confirmation that the SNP mutation is valid and carried by more than one man, the mutation is given a name something like R-M269 where R is the base haplogroup and M269 reflects the lab that discovered and named the SNP (M = Peter Underhill at Stanford) and an additional number, generally the next incremental number named by that lab (269).
Some SNPs were discovered simultaneously by different labs. When that happens, the same mutation in the identical location is given different names by different organizations, resulting in multiple names for the name mutation in the same DNA location. These are considered equivalent SNPs because they are identical.
In some cases, SNPs in different locations seem to define the same tree branching structure. These are functionally equivalent until enough tests are taken to determine a new branching structure, but they are not equivalent in the sense that the exact same DNA location was named by two different labs.
Some confusion exists about Y DNA SNP equivalence.
| Equivalence Confusion | How This Happens | Are They the Same? |
| Same exact DNA location named by two labs | Different SNP names for the same DNA location, named by two different labs at about the same time | Exactly equivalent because SNPs are named for the the exact same DNA locations, define only one tree branch ever |
| Different DNA locations and SNP names, one current tree branch | Different SNPs temporarily located on same branch of the tree because branches or branching structure have not yet been defined | When enough men test, different branches will likely be sorted out for the non-equivalent SNPs pointing to newly defined branch locations that divide the tree or branch |
Let’s look at an example where 4 example SNPs have been named. Two at the same location, and two more for two additional locations. However, initially, we don’t know how this tree actually looks, meaning what is the base/trunk and what are branches, so we need more tests to identify the actual structure.

The example structure of a haplogroup R branch, above, shows that there are three actual SNP locations that have been named. Location 1 has been given two different SNP names, but they are the same exact location. Duplicate names are not intentionally given, but result from multiple labs making simultaneous discoveries.
However, because we don’t have enough information yet, meaning not enough men have tested that carry at least some of the mutations (variants,), we can’t yet define trunks and branches. Until we do, all 4 SNPs will be grouped together. Examples 1 and 2 will always be equivalent because they are simply different names for the exact same DNA location. Eventually, a branching structure will emerge for Examples 1/2, Example 3 and Example 4..

Eventually, the downstream branches will be defined and split off. It’s also possible that Example 4 would be the trunk with Examples 1 and 2 forming a branch and Example 3 forming a branch. Branching tree structure can’t be built without sufficient testers who take the NGS tests, specifically the Big Y-700 which doesn’t just confirm a subset of existing named SNPs, but confirms all named SNPs, unnamed variants and discovers new previously-undiscovered variants which define the branching tree structure.
SNP testing occurs in multiple ways, including:
- NGS, next generation sequencing, tests such as the Big Y-700 which scans the gold standard region of the Y chromosome in order to find known SNPs at specific locations, mutations (variants) not yet named as SNPs, previously undiscovered variants and minimally 700 STR mutations.
- WGS, whole genome sequencing although there currently exist no bundled commercial tools to separate Y DNA information from the rest of the genome, nor any comparison methodology that allows whole genome information to be transferred to Family Tree DNA, the only commercial lab that does both testing and matching of NGS Y DNA tests and where most of the Y DNA tests reside. There can also be quality issues with whole genome sequencing if the genome is not scanned a similar number of times as the NGS Y tests. The criteria for what constitues a “positive call” for a mutation at a specific location varies as well, with little standardization within the industry.
- Targeted SNP testing of a specific SNP location. Available at Family Tree DNA and other labs for some SNP locations, this test would only be done if you are looking for something very specific and know what you are doing. In some cases, a tester will purchase one SNP to verify that they are in a particular lineage, but there is no benefit such as matching. Furthermore, matching on one SNP alone does not confirm a specific lineage. Not all SNPs are individually available for purchase. In fact, as more SNPs are discovered at an astronomical rate, most aren’t available to purchase separately.
- SNP panels which test a series of SNPs within a certain haplogroup in order to determine if a tester belongs to a specific subclade. These tests only test known SNPs and aren’t tests of discovery, scanning the useable portion of the Y chromosome. In other words, you will discern whether you are or are not a member of the specific subclades being tested for, but you will not learn anything more such as matching to a different subclade, or new, undiscovered variants (mutations) or subclades.
Subclade – A branch of a specific upstream branch of the haplotree.

For example, in haplogroup R, R1 and R2 are subclades of haplogroup R. The graphic above conveys the concept of a subclade. Haplogroups beneath R1 and R2, respectively, are also subclades of haplogroup R as well as subclades of all clades above them on the haplotree.
Older naming conventions used letter number conventions such as R1 and R2 which expanded to R1b1c and so forth, alternating letters and numbers.
Today, we see most haplogroups designated by the haplogroup letter and SNP name. Using that notation methodology, R would be R-M207, R1 would be R-M173 and R2 would be R-M479.

ISOGG documents Y haplogroup naming conventions and their history, maintaining both an alphanumeric and SNP tree for backwards compatibility. The reason that the alphanumeric tree was obsoleted was because there was no way to split a haplogroup like R1b1c when a new branch appeared between R1b and R1b1 without renaming everything downstream of R1b, causing constant reshuffling and renaming of tree branches. Haplogroup names were becoming in excess of 20 characters long. Today, the terminal SNP is used as a person’s haplogroup designation. The SNP name never changes and the individual’s Y haplogroup only changes if:
- Further testing is performed and the tester is discovered to have an additional mutation further downstream from their current terminal SNP
- A SNP previously discovered using the Big Y NGS test has since been named because enough men were subsequently discovered to carry that mutation, and the newly named SNP is the tester’s terminal SNP
Terminal SNP – It’s really not fatal. Used in this context, “terminal” means end of line, meaning furthest down and closest to present in the haplotree.
Depending on what level of testing you’ve undergone, you may have different haplogroups, or SNPs, assigned as your official “end of line” haplogroup or “terminal SNP” at various times.
If you took any of the various STR panel tests (12, 25, 37, 67 or 111) at Family Tree DNA your SNP was predicted based on STR matches to other men. Let’s say that prediction is R-M198. At that time, R-M198 was your terminal SNP. If you took the Big Y-700 test, your terminal SNP would almost assuredly change to something much further downstream in the haplotree.
If you took an autosomal test, your haplogroup was predicted based on a panel of SNPs selected to be informative about Y or mitochondrial DNA haplogroups. As with predicted haplogroups from STR test panels, the only way to discover a more definitive haplogroup is with further testing.
If you took a Y DNA STR test, you can see by looking at your match list that other testers may have a variety of “terminal SNPs.”

In the above example, the tester was originally predicted as R-M198 but subsequently took a Big Y test. His haplogroup now is R-YP729, a subclade of R-M198 several branches downstream.
Looking at his Y DNA STR matches to view the haplogroups of his matches, we see that the Y DNA predicted or confirmed haplogroup is displayed in the Y-DNA Haplogroup column – and several other men are M198 as well.
Anyone who has taken any type of confirming SNP test, whether it’s an individual SNP test, a panel test or the Big Y has their confirmed haplogroup at that level of testing listed in the Terminal SNP column. What we don’t know and can’t tell is whether the men whose Terminal SNP is listed as R-M198 just tested that SNP or have undergone additional SNP testing downstream and tested negative for other downstream SNPs. We can tell if they have taken the Big Y test by looking at their tests taken, shown by the red arrows above.
If the haplogroup has been confirmed by any form of SNP testing, then the confirmed haplogroup is displayed under the column, “Terminal SNP.” Unfortunately, none of this testers’ matches at this STR marker level have taken the Big Y test. As expected, no one matches him on his Terminal SNP, meaning his SNP farthest down on the tree. To obtain that level of resolution, one would have to take the Big Y test and his matches have not.
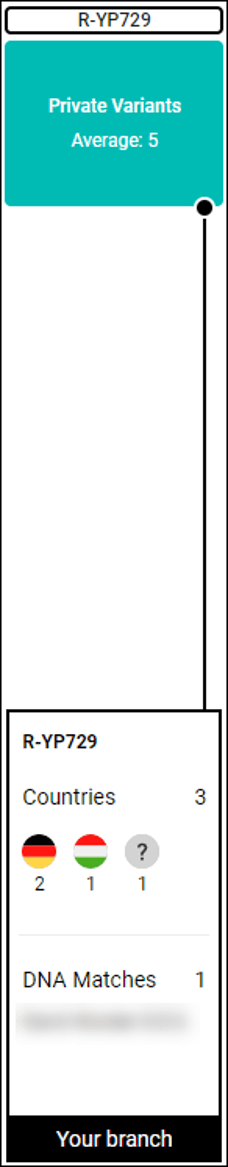
Looking at this tester’s Big Y Block Tree results, we can see that there are indeed 3 people that match him on his terminal SNP, but none of them match him on the STR tests which generally produce genealogical matches closer in time. This suggests that these haplogroup level matches are a result of an ancestor further back in time. Note that these men also have an average of 5 variants each that are currently unnamed. These may eventually be named and become baby branches.
SNP matches can be useful genealogically, depending on when they occurred, or can originate further back in time, perhaps before the advent of surnames.
Our tester’s paternal ancestors migrated from Germany to Hungary in the late 1700s or 1800s, settling in a region now in Croatia, but he’s brick-walled on his paternal line due to record loss during the various wars.
The block tree reveals that the tester’s Big Y SNP match is indeed from Germany, born in 1718, with other men carrying this same terminal SNP originating in both Hungary and Germany even though they aren’t shown as a STR marker match to our tester.
You can read more about the block tree in the article, Family Tree DNA’s New Big Y Block Tree.
Haplotype – your individual values for results of gene sequencing, such as SNPs or STR values tested in the 12, 25, 37, 67 and 111 marker panels at Family Tree DNA. The haplotype for the individual shown below would be 13 for location DYS393, 26 for location DYS390, 16 for location DYS19, and so forth.

The values in a haplotype tend to be inherited together, so they are “unique” to you and your family. In this case, the Y DNA STR values of 13, 26, 16 and 10 are generally inherited together (unless a new mutation occurs,) passed from father to son on the Y chromosome. Therefore, this person’s haplotype is 13, 26, 16 and 10 for these 4 markers.
If this haplotype is rare, it may be very unique to the family. If the haplotype is common, it may only be unique to a much larger haplogroup reaching back hundreds or thousands of years. The larger the haplotype, the more unique it tends to be.
STR – Short tandem repeat. I think of a short tandem repeat as a copy machine or a stutter error. On the Y chromosome, the value of 13 at the location DYS393 above indicates that a series of DNA nucleotides is repeated a total of 13 times.
![]()
Starting with the above example, let’s see how STR values accrue mutations.
![]()
In the example above, the value of CT was repeated 4 times in this DNA sequence, for a total of 5, so 5 would be the marker value.
![]()
DNA can have deletions where the DNA at one or more locations is deleted and no DNA is found at that location, like the missing A above.
DNA can also have insertions where a particular value is inserted one or more times.
![]()
For example, if we know to expect the above values at DNA locations 1-10, and an insertion occurs between location 3 and 4, we know that insertion occurred because the alignment of the pattern of values expected in locations 4-10 is off by 1, and an unexpected T is found between 3 and 4, which I’ve labeled 3.1.
![]()
STR, or copy mutations are different from insertions, deletions or SNP mutations, shown below, where one SNP value is actually changed to another nucleotide.
![]()
Haplotree – the SNP trees of humanity. Just a few years ago, we thought that there were only a few branches on the Y and mitochondrial trees of humanity, but the Big Y test has been a game changer for Y DNA.
At the end of 2019, the tree originating in Africa with Y chromosome Adam whose descendants populated the earth is comprised of more than 217,277 variants divided into 24,838 individual Y haplotree branches
A tree this size is very difficult to visualize, but you can take a look at Family Tree DNA’s public Y DNA tree here, beginning with haplogroup A. Today, there 25,880 branches, increased by more than 1000 branches in less than 3 weeks since year end. This tree is growing at breakneck speed as more men take the Big Y-700 test and new SNPs are discovered.
On the Public Y Tree below, as you expand each haplogroup into subgroups, you’ll see the flags representing the locations of where the testers’ most distant paternal ancestor lived.

I wrote about how to use the Y tree in the article Family Tree DNA’s PUBLIC Y DNA Haplotree.
The mitochondrial tree can be viewed here. I wrote about to use the mitochondrial tree in the article Family Tree DNA’s Mitochondrial Haplotree.
Need Something Else?
I’ll be introducing more concepts and terms in future articles on the various Y DNA features. In the mean time, be sure to use the search box located in the upper right-hand corner of the blog to search for any term.

For example, want to know what Genetic Distance means for either Y or mitochondrial DNA? Just type “genetic distance” into the search box, minus the quote marks, and press enter.
Enjoy and stay tuned for Part 3 in the Y DNA series, coming soon.
______________________________________________________________
Sign Up Now – It’s Free!
If you enjoyed this article, subscribe to DNAeXplain for free, to automatically receive new articles by emailed each week.
Here’s the link. Just look for the little grey “follow” button on the right-hand side on your computer screen below the black title bar, enter your e-mail address, and you’re good to go!
In case you were wondering, I never have nor ever will share or use your e-mail outside of the intended purpose.
Share the Love
You can always forward these articles to friends or share by posting links on social media. Who do you know that might be interested?
_____________________________________________________________
Disclosure
I receive a small contribution when you click on some of the links to vendors in my articles. This does NOT increase the price you pay but helps me to keep the lights on and this informational blog free for everyone. Please click on the links in the articles or to the vendors below if you are purchasing products or DNA testing.
Thank you so much.
DNA Purchases and Free Transfers
- FamilyTreeDNA – Y, mitochondrial and autosomal DNA testing
- MyHeritage DNA – ancestry autosomal DNA only, not health
- MyHeritage DNA plus Health
- MyHeritage FREE DNA file upload – transfer your results from other vendors free
- AncestryDNA – autosomal DNA only
- 23andMe Ancestry – autosomal DNA only, no Health
- 23andMe Ancestry Plus Health
- LivingDNA
Genealogy Products and Services
- MyHeritage FREE Tree Builder – genealogy software for your computer
- MyHeritage Subscription with Free Trial
- Legacy Family Tree Webinars – genealogy and DNA classes, subscription based, some free
- Legacy Family Tree Software – genealogy software for your computer
- Charting Companion – Charts and Reports to use with your genealogy software or FamilySearch
Genealogy Research
- Legacy Tree Genealogists – professional genealogy research
Fun DNA Stuff
- Celebrate DNA – customized DNA themed t-shirts, bags and other items


Dear Roberta.
Many thanks for the article on nomenclature. I hope that people writing articles or comments om DNA in genealogy will follow the scientific nomenclature and not their more or less private nomenclature.
I have one further comment on your article: The double helix you present is not DNA. The DNA helix is right-twisted, like a screw in your toolbox. The double helix you present in the article is left twisted. You will not find this DNA in nature.
Finally, you and most writers use blue, green and red colors to characterize segregation of chromosomes or segments of DNA. Eight percent of male readers including me, have impaired red-green color vision and consequently cannot follow your arguments. Please use blue, yellow and brown instead.
All the best, CB. Oslo.
Regarding the helix, I was simply trying to find a colorized version of a helix with nucleotides that was not copyrighted that I could use for the article. I am not a graphics artist nor expert. The helix “rungs” need 4 colors, not three, so do you have another color in addition to blue, yellow and brown that you can see? Also, do you have the graphics expertise to create a helix with appropriately colored and placed nucleotides? Thanks.
Dear Roberta.
I fully understand your troubles in finding a not copyrighted version of DNA. Maybe I can help you. One of my sons, Jakob, (in data busyness) is also a skilled graphic writer. I will ask him, and if you like the results, you alone will be free to use it.
I suggest the following criteria: 1. considerably shorter length of DNA than the one you used, 2. right-twisted (!), 3. showing the short thymine T black, the large adenine A white, the short cytosine C brown, the large guanine G yellow.
I realize that FTDNA and others use red/green colors in presenting chromosome materials, DNA browser for instance, using red/green variants that I (and 8 % of males) do not see. I still fight for our right to be presented comprehensible information.
CBvdH
Thank you. I appreciate the help.
I thought I’d have a go at this illusttration, but the 1st thing I noticed .. is that the “twist” is ambiguous, meaning dependent on which strand you’re following and from which end! i.e. one strand is right twist while the other is left twist. Color is another mater of course!
Roberta, thanks for Part 2 regarding Y-DNA. In the Haplogroup discussion you state “I’ve as many as two mutations in a father son pair,…” This would be extremely rare if I understand the theory correctly. Can you provide a little more detail on how many father-son pairs with more than one mutation difference that you have seen? Thank you.
I haven’t kept track, or looked for them. I can think of 2 cases off the top of my head though.
Nice follow-up to article one. Still anxiously awaiting three though as I’m debating the merits of uploading my new Big Y-700 data to YFull using only the VCF file at 1st, or waiting until I can afford to buy my BAM file. I want to understand the -practical-relationship between SNPs & STRs as it applies to lineage verification.
On the topic of terminology, I agree with the 1st comment but since most everyone is using FTDNA I think you should note that additional SNP “panels” are called “SNP Packs” when they are put together to narrow down a haplogroup and (hopefully) produce a terminal SNP with a standard marker STR test result.
You mention that a haplogroup is confirmed by FTDNA when an STR prediction has been confirmed by SNP testing, however in looking at my Y-DNA match list (std, not Big-Y) I see that one match who I don’t believe has tested with more than the 111 marker test is assigned a terminal SNP. As I recall myself and one other match were also assigned this terminal SNP at 111 markers, but the two of us did an SNP Pack and were reassigned a rare downstream terminal SNP on the branch. I don’t have screen caps prior to us taking the SNP Pack so I can’t say for certain that we were all listed with this upstream Terminal SNP. Is it possible that because of our SNP Pack and my Big-Y testing that now this other match may have had the downstream Terminal SNP assigned from previously being just listed as the haplogroup (prediction)? (Hope this makes sense!)
I am not addressing YFull and the VCF/BAM question.
Ok. I’ve asked FTDNA and you are correct (I never doubted, only was unclear about the FTDNA display & my memory) that a “Terminal SNP” display indicates SNP testing, but if done via an add-on panel (SNP Pack) the display does not show that.
Which means that even if the haplogroup “prediction” is VERY accurate (as it was with my 2 matches) it is not a guarantee of that being final until confirmed by SNP testing. What confused me was that for about a year now one match remained at the same upstream haplogroup designation (as I and my other match was until a SNP Pack), but apparently he has since done a SNP Pack that actually confirmed it as his terminal SNP, but the only way to know was that it is now displayed as “Terminal SNP”.
So for us, we are all the same somewhat unusual “lines” but have branched about 2,500 years ago. I think I’m clear on the subtle display differences now.
.. and the answer to my last question (last sentence) is- No.
Thank you Roberta. Very timely, as I am bogged down in writing a “simple” Y-DNA to BIg Y primer for our surname project. Now, I can simply point to your article for a more in-depth discussion.
1. “Some SNPs were discovered simultaneously by different labs.”
True but in the past these were more often FTDNA tests “interpreted’ by others, and not tested. With FTDNA starting to name with BY prefixed names for Big Y-500, and now FT prefixes for Big Y-700 SNPs, we will see these start to dominate (if not already).
https://isogg.org/wiki/FT_SNP_index
2. “Some confusion exists about Y DNA SNP equivalence”
Not true, the confusion only arises because of incorrect terminology.
Phylogenetically Equivalent: A term used when describing the relationship between two or more SNPs; specifically, SNPs that belong on the same branch (or clade) of a haplogroup or phylogenetic tree, for example, Y-DNA haplogroup M is defined by the following SNPs: M4, M5, M106, M186, M189, P35, which are said to be phylogenetically equivalent.
https://isogg.org/tree/ISOGG_Glossary.html
Equivalent .. describing persons (read SNPs) who were equal in power or rank (read age)
https://www.lexico.com/definition/equivalent
Synonymous ……. having the same meaning as another word
https://www.lexico.com/definition/synonymous
3. Panel test vs. SNP Pack
FTDNA use the term panel in respect of STRs, so Y1-12 is Panel 1, but YSeq uses the term panel for both STRs and SNPs. FTDNA SNP Packs are carefully selected groups of SNPs, eg. R1b Backbone SNP Pack.
4. Colours are important for all of us. I do hope taht the liaison with Carl helps.
When my first cousin was tested at 67 markers he was initially assigned to J-M172 with a terminal SNP of PF5456. I joined his kit to the J-M172 project and after talking with the managers there they suggested further (STR?) testing the details of which I don’t recall any more. The result now is that his “Confirmed Halplogroup” is J-BY268. I do not see a BY268 on his match page although he does have one PF5456. However it is at GD 7 so I don’t know if it is really telling me anything useful. He also has a match at GD 6 which is identified as FGC21083 which I note is just below his assigned haplogroup in his tree. His closest match, and everything above GD 5, don’t show terminal SNPS and he has no surname matches. His GD 2 match never answered my email.
So is there anything useful in all of this that suggests what I should do now?
Roberta,
Out of curiosity, have you heard anything back from FTDNA lately about those y-DNA D0 kits that were uploaded last summer, shortly after the D0 paper from Marc Haber came out? Looking at the D tree on FTDNA, there are two basal kits under D-FT75 (one Syrian, one American), an American under D-FT76, and two Saudis under D-FT155. I’m curious if you know the ethnic background of the American kits? I suspect one of them is African American, just not sure which.
https://www.familytreedna.com/public/y-dna-haplotree/D
Do you know if they have any other D0 kits waiting to be analyzed?
I don’t know, I’m sorry.
Regardless of where the SNP names come from per John Templar Harper’s post, the confusion that Roberta mentions about the term “equivalent SNPs” is not common among at least the community of citizen-scientists who have worked with Y-DNA for years.
Usually “synonym SNPs” is used when there are two (or more) names for the same physical mutation at the same position. Ex: DF1 and L513 are synonyms or synonym SNPs. Sounds like that terminology hasn’t been well-distributed and should be better advertised.
“Equivalent SNPs” is used when there are two or more SNPs within a block (also called an “equivalent block”) on the haplotree that hasn’t been split up yet by additional downstream testers.
Synonyms (as in synonym SNPs) are synonyms forever. Equivalents are only equivalent for as long as they remain in the same block, once they’re broken into separate blocks they’re no longer equivalents.
Pingback: Genetic Genealogy at 20 Years: Where Have We Been, Where Are We Going and What’s Important? | DNAeXplained – Genetic Genealogy