Beginning with the full sequencing of the Neanderthal genome, first published in May 2010 by the Max Planck Institute with Svante Paabo at the helm, and followed shortly thereafter with a Denisovan specimen, we began to unravel our ancient history.
The photo below shows a step in the process of extracting DNA from ancient bones at Max Planck.
Our Y and mitochondrial DNA haplogroups take us back thousands of years in time, but at some point, where and how people were settling and intermixing becomes fuzzy. Ancient DNA can put the people of that time and place in context. We have discovered that current populations do not necessarily represent the ancient populations of a particular locale.
Recent information discovered from ancient burials tells us that the people of Europe descend from a 3 pronged model. Until recently, it was believed that Europeans descended from Paleolithic hunter-gatherers and Neolithic farmers, a two-pronged model.
Previously, it was believed that Europe was peopled by the ancient hunter-gatherers, the Paleolithic, who originally settled in Europe beginning about 45,000 years ago. At this time, the Neanderthal were already settled in Europe but weren’t considered to be anatomically modern humans, and it was believed, incorrectly, that the two groups did not interbreed. These hunter-gatherers were the people who settled in Europe before the last major ice age, the Younger Dryas, taking refuge in the southern portions of Europe and Eurasia, and repeopling the continent after the ice receded, about 12,000 years ago. By that time, the Neanderthals were gone, or as we now know, at least partially assimilated.
This graphic shows Europe during the last ice age.
The second settlement wave, the agriculturalist farmers from the Near East either overran or integrated with the hunter-gatherers in the Neolithic period, depending on which theory you subscribe to, about 8000-10,000 years ago.
2012 – Ancient Northern European (ANE) Hints
Beginning in 2012, we began to see hints of a third lineage that contributed to the peopling of Europe as well, from the north. Buried in the 2012 paper, Estimating admixture proportions and dates with ADMIXTOOLS by Patterson et al, was a very interesting tidbit. This new technique showed a third population, referred to by many as a “ghost population”, because no one knew who they were, that contributed to the European population.
The new population was termed Ancient North Eurasian, or ANE.
Dienekes covered this paper in his blog, but without additional information, in the community in general, there wasn’t much more than a yawn.
2013 – Mal’ta Child Stirs Excitement
The first real hint of meat on the bones of ANE came in the form of ancient DNA analysis of a 24,000 year old Siberian boy that has come to be named Mal’ta (Malta) Child. In the original paper, by Raghaven et al, Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans, he was referred to as MA-1. I wrote about this in my article titled Native American Gene Flow – Europe?, Asia and the Americas. Dienekes wrote about this paper as well.
This revelation caused quite a stir, because it was reported that the Ancestor of Native Americans in Asia was 30% Western Eurasian. Unfortunately, in some cases, this was immediately interpreted to mean that Native Americans had come directly from Europe which is not what this paper said, nor inferred. It was also inferred that the haplogroups of this child, R* (Y) and U (mtDNA) were Native American, which is also incorrect. To date, there is no evidence for migration to the New World from Europe in ancient times, but that doesn’t mean we aren’t still looking for that evidence in early burials.
What this paper did show was that Europeans and Native Americans shared a common ancestor, and that the Siberian population had contributed to the European population as well as the Native American population. In other words, descendants settled in both directions, east and west.
The most fascinating aspect of this paper was the match distribution map, below, showing which populations Malta child matched most closely.
As you can see, MA-1, Malta Child, matches the Native American population most closely, followed by the northern European and Greenland populations. The further south in Europe and Asia, the more distant the matches and the darker the blue.
2013 – Michael Hammer and Haplogroup R
Last fall at the Family Tree DNA conference, Dr. Michael Hammer, from the Hammer Lab at the University of Arizona discussed new findings relative to ancient burials, specifically in relation to haplogroup R, or more specifically, the absence of haplogroup R in those early burials.
Based on the various theories and questions, ancient burials were enlightening.
In 2013, there were a total of 32 burials from the Neolithic period, after farmers arrived from the Near East, and haplogroup R did not appear. Instead, haplogroups G, I and E were found.
What this tells us is that haplogroup R, as well as other haplogroup, weren’t present in Europe at this time. Having said this, these burials were in only 4 locations and, although unlikely, R could be found in other locations.
Last year, Dr. Hammer concluded that haplogroup R was not found in the Paleolithic and likely arrived with the Neolithic farmers. That shook the community, as it had been widely believed that haplogroup R was one of the founding European haplogroups.
While this provided tantalizing information, we still needed additional evidence. No paper has yet been published that addresses these findings. The mass full sequencing of the Y chromosome over this past year with the introduction of the Big Y will provide extremely valuable information about the Y chromosome and eventually, the migration path into and across Europe.
2014 – Europe’s Three Ancient Tribes
In September 2014, another paper was published by Lazaridis et al that more fully defined this new ANE branch of the European human family tree. An article in BBC News titled Europeans drawn from three ancient ‘tribes’ describes it well for the non-scientist. Of particular interest in this article is the artistic rendering of the ancient individual, based on their genetic markers. You’ll note that they had dark skin, dark hair and blue eyes, a rather unexpected finding.
In discussing the paper, David Reich from Harvard, one of the co-authors, said, “Prior to this paper, the models we had for European ancestry were two-way mixtures. We show that there are three groups. This also explains the recently discovered genetic connection between Europeans and Native Americans. The same Ancient North Eurasian group contributed to both of them.”
The paper, Ancient human genomes suggest three ancestral populations for present-day Europeans, appeared as a letter in Nature and is behind a paywall, but the supplemental information is free.
The article summary states the following:
We sequenced the genomes of a ~7,000-year-old farmer from Germany and eight ~8,000-year-old hunter-gatherers from Luxembourg and Sweden. We analysed these and other ancient genomes1, 2, 3, 4 with 2,345 contemporary humans to show that most present-day Europeans derive from at least three highly differentiated populations: west European hunter-gatherers, who contributed ancestry to all Europeans but not to Near Easterners; ancient north Eurasians related to Upper Palaeolithic Siberians3, who contributed to both Europeans and Near Easterners; and early European farmers, who were mainly of Near Eastern origin but also harboured west European hunter-gatherer related ancestry. We model these populations’ deep relationships and show that early European farmers had ~44% ancestry from a ‘basal Eurasian’ population that split before the diversification of other non-African lineages.
This paper utilized ancient DNA from several sites and composed the following genetic contribution diagram that models the relationship of European to non-European populations.
Present day samples are colored purple, ancient in red and reconstructed ancestral populations in green. Solid lines represent descent without admixture and dashed lines represent admixture. WHG=western European hunter-gatherer, EEF=early European farmer and ANE=ancient north Eurasian
2014 – Michael Hammer on Europe’s Ancestral Population
For anyone interested in ancient DNA, 2014 has been a banner years. At the Family Tree DNA conference in Houston, Texas, Dr. Michael Hammer brought the audience up to date on Europe’s ancestral population, including the newly sequenced ancient burials and the information they are providing.
Dr. Hammer said that ancient DNA is the key to understanding the historical processes that led up to the modern. He stressed that we need to be careful inferring that the current DNA pattern is reflective of the past because so many layers of culture have occurred between then and now.
Until recently, it was assumed that the genes of the Neolithic farmers replaced those of the Paleolithic hunter-gatherers. Ancient DNA is suggesting that this is not true, at least not on a wholesale level.
The theory, of course, is that we should be able to see them today if they still exist. The migration and settlement pattern in the slide below was from the theory set forth in the 1990s.
In 2013, Dr. Hammer discussed the theory that haplogroup R1b spread into Europe with the farmers from the Near East in the Neolithic. This year, he expanded upon that topic that based on the new findings from ancient burials.
Last year, Dr. Hammer discussed 32 burials from 4 sites. Today, we have information from 15 ancient DNA sites and many of those remains have been full genome sequenced.
Information from papers and recent research suggests that Europeans also have genes from a third source lineage, nicknamed the “ghost population of North Eurasia.”
Scientists are finding a signal of northeast Asian related admixture in northern Europeans, first suggested in 2012. This was confirmed with the sequencing of Malta child and then in a second sequencing of Afontova Gora2 in south central Siberia.
We have complete genomes from nine ancient Europeans – Mesolithic hunter gatherers and Neothilic farmers. Hammer refers to the Mesolithic here, which is a time period between the Paleolithic (hunter gatherers with stone tools) and the Neolithic (farmers).
In the PCA charts, shown above, you can see that Europeans and people from the Near East cluster separately, except for a bridge formed by a few Mediterranean and Jewish populations. On the slide below, the hunter-gatherers (WHG) and early farmers (EEF) have been overlayed onto the contemporary populations along with the MA-1 (Malta Child) and AG2 (Afontova Gora2) representing the ANE.
When sequenced, separate groups formed including western hunter gathers and early european farmers include Otzi, the iceman. A third group is the north south clinal variation with ANE contributing to northern European ancestry. The groups are represented by the circles, above.
Dr. Hammer said that the team who wrote the “Ancient Human Genomes” paper just recently published used an F3 test, results shown above, which shows whether populations are an admixture of a reference population based on their entire genome. He mentioned that this technique goes well beyond PCA.
Mapped onto populations today, most European populations are a combination of the three early groups. However, the ANE is not found in the ancient Paleolithic or Neolithic burials. It doesn’t arrive until later.
This tells us that there was a migration event 45,000 years ago from the Levant, followed about 7000 years ago by farmers from the Near East, and that ANE entered the population some time after that. All Europeans today carry some amount of ANE, but ancient burials do not.
These burials also show that southern Europe has more Neolithic farmer genes and northern Europe has more Paleolithic/Mesolithic hunter-gatherer genes.
Pigmentation for light skin came with farmers – blue eyes existed in hunter gatherers even though their skin was dark.
Dr. Hammer created these pie charts of the Y and mitochondrial haplogroups found in the ancient burials as compared to contemporary European haplogroups.
The pie chart on the left shows the haplogroups of the Mesolithic burials, all haplogroup I2 and subclades. Note that in the current German population today, no I2a1b and no I1 was found. The chart on the right shows current Germans where haplogroup I is a minority.
Therefore, we can conclude that haplogroup I is a good candidate to be identified as a Paleolithic/Mesolithic haplogroup.
This information shows that the past is very different from today.
In 2014 we have many more burials that have been sequenced than last year, as shown on the map above.
Green represents Neolithic farmers, red are Mesolithic hunter-gatherers, brown at bottom right represents more recent samples from the Metallic age.
There are a total of 48 Neolithic burials where haplogroup G dominates. In the Mesolithic, there are a total of six haplogroup I.
This suggests that haplogroup I is a good candidate to be the father of the Paleolithic/Mesolithic and haplogroup G, the founding father of the Neolithic.
In addition to haplogroup G in the Neolithic, one sample of both E1b1b1 (M35) and C were also found in Spain. E1b1b1 isn’t surprising given it’s north African genesis, but C was quite interesting.
The Metal ages, which according to wiki begin about 3300BC in Europe, is where haplogroup R, along with I1, first appear.
Please note that the diffusion of melallurgy map above is not part of Dr. Hammer’s presentation. I have added it for clarification.
Nothing is constant in Europe. The Y DNA was very upheaved, as indicated on the graphic above. Mitochondrial DNA shifted from pre-Neolithic to Neolithic which isn’t terribly different from the present day.
Dr. Hammer did not say this, but looking at the Y versus the mtDNA haplogroups, I wonder if this suggests that indeed there was more of a replacement of the males in the population, but that the females were more widely assimilated. This would certainly make sense, especially if the invaders were warriors and didn’t have females with them. They would have taken partners from the invaded population.
Haplogroup G represents the spread of farming into Europe.
The most surprising revelation is that haplogroup R1b appears to have emerged after the Neolithic agriculture transition. Given that just three years ago we thought that haplogroup R1b was one of the original European settlers thousands of years ago, based on the prevalence of haplogroup R in Europe today, at about 50%, this is a surprising turn of events. Last year’s revelation that R was maybe only 7000-8000 years old in Europe was a bit of a whammy, but the age of R in Europe in essence just got halved again and the source of R1b changed from the Near East to the Asian steppes.
Obviously, something conferred an advantage to these R1b men. Given that they arrived in the early Metalic age, was it weapons and chariots that enabled the R1b men who arrived to quickly become more than half of the population?
The Bronze Age saw the first use of metal to create weapons. Warrior identity became a standard part of daily life. Celts ranged over Europe and were the most dominant iron age warriors. Indo-European languages and chariots arrived from Asia about this time.
The map above shows the Hallstadt and LaTene Celtic cultures in Europe, about 600BC. This was not a slide presented by Dr. Hammer.
Haplogroup R1b was not found in an ancient European context prior to a Bell Beaker period burial in Germany 4.8-4.0 kya (thousand years ago, i.e. 4,800-4,000 years ago). R1b arrives about 4.6 kya and is also found in a Corded Ware culture burial in Germany. A late introduction of these lineages which now predominate in Europe corresponds to the autosomal signal of the entry of Asian and Eastern European steppe invaders into western Europe.
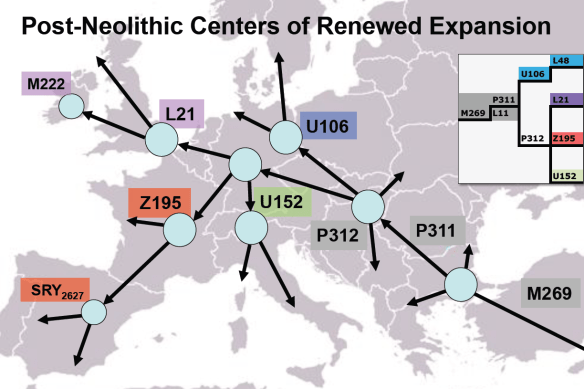
Local expansion occurred in Europe of R1b subgroups U106, L21 and U152.
A current haplogroup R distribution map that reflects the findings of this past year is shown above.
Haplogroup I is interesting for another reason. It looks like haplogroup I2a1b (M423) may have been replaced by I1 which expanded after the Mesolithic.
On the slide above, the Loschbour sample from Luxembourg was mapped onto a current haplogroup I SNP map where his closest match is a current day Russian.
One of the benefits of ancient DNA genome processing is that we will be able to map current trees into maps of old SNPs and be able to tell who we match most closely.
Autosomal DNA can also be mapped to see how much of our DNA is from which ancient population.
Dr. Hammer mapped the percentages of European Mesolithic/Paleolithic hunter-gatherers in blue, Neolithic Farmers from the Near East in magenta and Asian Steppe Invaders representing ANE in yellow, over current populations. Note the ancient DNA samples at the top of the list. None of the burials except for Malta Child carry any yellow, indicating that the ANE entered the European population with the steppe invaders; the same group that brought us haplogroup R and possibly I1.
Dr. Hammer says that ANE was introduced to and assimilated into the European population by one or more incursions. We don’t know today if ANE in Europeans is a result of a single blast event or multiple events. He would like to do some model simulations and see if it is related to timing and arrival of swords and chariots.
We know too that there are more recent incursions, because we’re still missing major haplogroups like J.
The further east you go, meaning the closer to the steppes and Volga region, the less well this fits the known models. In other words, we still don’t have the whole story.
At the end of the presentation, Michael was asked if the whole genomes sequenced are also obtaining Y STR data, which would allow us to compare our results on an individual versus a haplogroup level. He said he didn’t know, but he would check.
Family Tree DNA was asked if they could show a personal ancient DNA map in myOrigins, perhaps as an alternate view. Bennett took a vote and that seemed pretty popular, which he interpreted as a yes, we’d like to see that.
In Summary
The advent of and subsequent drop in the price of whole genome sequencing combined with the ability to extract ancient DNA and piece it back together have provided us with wonderful opportunities. I think this is jut the proverbial tip of the iceberg, and I can’t wait to learn more.
If you are interested in other articles I’ve written about ancient DNA, check out these links:
- 400,000 Year Old DNA from Spain Sequenced
- Finding Your Inner Neanderthal with Evolutionary Geneticist Svante Paabo
- Neanderthal Genome Further Identified in Contemporary Eurasians
- Sequencing of Neanderthal Toe Bone Reveals Unknown Hominin Line
- Decoding and Rethinking Neanderthals
- DNA Analysis of 8000 Year Old Bones Allow Peek Into the Neolithic
- Native American Gene Flow – Europe?, Asia and the Americas
- Native Americans, Neanderthal and Denisova Admixture
- 5,500 Year Old Grandmother Found Using DNA
- Ancient DNA Analysis from Canada
- The Malhi Molecular Anthropology and Ancient DNA Labs
- Analyzing the Native American Clovis Anzick Ancient Results
- Utilizing Ancient DNA at GedMatch
- More Ancient DNA Samples for Comparison
- Ancient DNA Matches – What Do They Mean?
- Ancient DNA Matching – A Cautionary Tale
______________________________________________________________
Disclosure
I receive a small contribution when you click on some of the links to vendors in my articles. This does NOT increase the price you pay but helps me to keep the lights on and this informational blog free for everyone. Please click on the links in the articles or to the vendors below if you are purchasing products or DNA testing.
Thank you so much.
DNA Purchases and Free Transfers
- Family Tree DNA
- MyHeritage DNA only
- MyHeritage DNA plus Health
- MyHeritage FREE DNA file upload
- AncestryDNA
- 23andMe Ancestry
- 23andMe Ancestry Plus Health
- LivingDNA
Genealogy Services
Genealogy Research
- Legacy Tree Genealogists for genealogy research
Discover more from DNAeXplained - Genetic Genealogy
Subscribe to get the latest posts sent to your email.





















































Roberta, that is awesome. When are you going to put everything into a book? Do you have a publisher? What are the short list names for the book? Let us help you name the book.
It will be a great best seller!!
Thanks, but no book, no publisher and no time:) I discovered a couple years ago that you have to hire an agent and no agent wants to work with you unless you’re already proven. That’s when I decided to blog instead. It’s much for flexible too.
My brother-in-law is part Indian and hasn’t done a DNA test which test would be best to find what tribe he belong to?
Tom James
tjames@adcomworldwide.com
Generally, tribe can’t be determined. However, here is an article detailing the testing and what it can tell you. http://dna-explained.com/2012/12/18/proving-native-american-ancestry-using-dna/
Merci, Roberta. I feel as if I’ve been in a favourite class – but I’m running to catch up!! So much to learn, and to keep it in mind as I read the slides and your commentary/explanations. Fantastic. I appreciate all the work you’ve done here for the genealogy community.
“The most surprising revelation is that haplogroup R1b appears to have emerged after the Neolithic agriculture transition. Given that just three years ago we thought that haplogroup R1b was one of the original European settlers thousands of years ago, based on the prevalence of haplogroup R in Europe today, at about 50%, this is a surprising turn of events. Last year’s revelation that R was maybe only 7000-8000 years old in Europe was a bit of a whammy, but the age of R in Europe in essence just got halved again and the source of R1b changed from the Near East to the Asian steppes.”
This was actually something that had been accepted by many before three years ago. For example just look at the 2010 ISOGG Hg R page. The difference is the source being southwest Asia vs. Eurasian steppe.
“Haplogroup R1b1b2 (M269) is observed most frequently in Europe, especially western Europe, but with notable frequency in southwest Asia. R1b1b2 is estimated to have arisen approximately 4,000 to 8,000 years ago in southwest Asia and to have spread into Europe from there. The Atlantic Modal Haplotype, or AMH, is the most common STR haplotype in haplogroup R1b1b2a1a (P310/S129) and most European R1b1b2 belongs to haplogroups R1b1b2a1a1 (U106) or R1b1b2a1a2 (P312/S116).”
http://www.isogg.org/tree/ISOGG_HapgrpR10.html
At the same time if you had been keeping up with Jean Manco’s posts on various forums you wouldn’t have been so surprised. Jean Manco also published an excellent book Ancestral Journeys: The Peopling of Europe from the First Venturers to the Vikings which was published before Dr. Michael Hammer’s 2013 presentation.
Some of the slides used in Dr. Michael Hammer’s 2013 presentation are from Maciamo Hay’s Eupedia site where he had already provided the hypothesis of R1b going into western Europe in the post-Neolithic. http://www.eupedia.com/europe/Haplogroup_R1b_Y-DNA.shtml
It’s about time that everyone else such as Dr. Michael Hammer and Spencer Wells are getting on board although there are still some hold outs that don’t want to believe that R1b wasn’t in western Europe in the Mesolithic.
Apropos just published article from “The New York Times” on a paper from “Nature”:
http://www.nytimes.com/2014/10/23/science/research-humans-interbred-with-neanderthals.html?_r=0
And Dienekes’ coverage of the “Nature” paper:
http://dienekes.blogspot.com/2014/10/high-coverage-genome-from-45000-year.html
“The Metal ages, which according to wiki begin about 3300BC in Europe, is where haplogroup R, along with I1, first appear….”
If I’m not mistaken, one of the two samples from an LBK sites (6th millennium BC) from the Carpathian Basin belonged to I1 and the other belonged to G2a2b.
Loschbour and Stuttgart have been uploaded to FTDNA.
http://www.fc.id.au/2014/10/ancient-dna-matches-in-familytreedna.html
“In addition to haplogroup G in the Neolithic, one sample of both E1b1b1 (M35) and C were also found in Spain. E1b1b1 isn’t surprising given it’s north African genesis, but C was quite interesting.”
When I initially read that I figured you were talking about something other than the Avellaner cave specimens. The E1b1b1 specimen tested positive for V13 which has it’s highest frequency in the Balkans and all people that have it now are in Europe as far as I can tell. That E1b1b1 V13 was not directly from north Africa and the terminal SNP was not M35. The original study that reported that find and the DNA results can be seen at http://www.pnas.org/content/108/45/18255.full
I don’t believe that Dr. Hammer said where those specimens were from. Since C was not found in the Avellaner cave, there is at least one more source someplace.
Here’s the C sample. http://eurogenes.blogspot.com/2014/01/mesolithic-genome-from-spain-reveals.html
Yes,I was aware of the study. The Mesolithic Hunter Gatherer C specimen was from La Braña. It’s unfortunate that Dr. Hammer didn’t specify which study the Late Neolithic G and E-V13 specimens belonged to and which study the Mesolithic Hunter Gatherer C specimen belonged to although he did have the following two slides, that you posted, that did have some information on them although the Y-DNA info is not included. https://dnaexplained.files.wordpress.com/2014/10/hammer-2014-19.jpg and https://dnaexplained.files.wordpress.com/2014/10/hammer-2014-16.jpg
Dienekes post about the Late Neolithic G and E-V13 specimens can be found at http://dienekes.blogspot.com/2011/11/y-haplogroups-e-v13-and-g2a-in.html
He has the following statements –
“While this Neolithic E-V13 may well have come from the Balkans, and the common ancestor of the very uniform present-day Balkan cluster may have lived after this Spanish find, it is now certain that E-V13 was established in Europe long before the Bronze Age. This highlights the need to avoid Y-STR based calculations on modern populations for inferring patterns of ancient history, and not to conflate TMRCAs with “dates of arrival”: “In short: a particular TMRCA is consistent with either the arrival of the lineage long before and long after the TMRCA in a particular geographical area.”
“Analysis of shared haplotypes showed that the G2a haplotype found in ancient specimens is rare in current populations: its frequency is less than 0.3%(Table S3). The haplotype of individual ave07 is more frequent (2.44%), particularly in southeastern European populations (up to 7%). The Ave07 haplotype was also compared with current Eb1b1a2 haplotypes previously published (10–14). It appeared identical at the seven markers tested to five Albanian, two Bosnian, one Greek, one Italian, one Sicilian, two Corsican, and two Provence French samples and are thus placed on the same node of the E1b1b1a1b-V13 network as eastern, central, and western Mediterranean haplotypes (Fig. S1).
Pingback: Kostenki14 – A New Ancient Siberian DNA Sample | DNAeXplained – Genetic Genealogy
Pingback: Kostenki14 – A New Ancient Siberian DNA Sample | Native Heritage Project
When, where does Haplogroup T occur?
That here => https://dnaexplained.files.wordpress.com/2014/10/hammer-2013-11.png
is not the Global R1* distribution map. It is the global distrbution map of R1b.
This here is the global distrbution map of R1*
http://cache.eupedia.com/images/content/Haplogroup_R-borders.gif
Bad mistake guys. otherwise good work.
Pingback: 2014 Top Genetic Genealogy Happenings – A Baker’s Dozen +1 | DNAeXplained – Genetic Genealogy
Still searching and waiting for more info to be discovered on haplogroup Mt-DNA J2b1a.
My wife and her sister are J2b1a, and they live in Sweden and as far as we know come from a mostly Swedish but also to a smaller portion a Norwegian family that goes back hundreds of years.
christer@amneus.se
Pingback: Yamnaya, Light Skinned, Brown Eyed….Ancestors??? | DNAeXplained – Genetic Genealogy
Pingback: DNAeXplain Archives – General Information Articles | DNAeXplained – Genetic Genealogy
Roberta
Nice summary , thanks
Can you read the fine print as to the ID of the ? HDP russian who closest match to Loschbour ?
Pingback: Ethnicity Testing and Results | DNAeXplained – Genetic Genealogy
Pingback: Native American Haplogroup X2a – Solutrean, Hebrew or Beringian? | DNAeXplained – Genetic Genealogy
Pingback: Native American Haplogroup X2a – Solutrean, Hebrew or Beringian? | Native Heritage Project
Pingback: Ethnicity Testing – A Conundrum | DNAeXplained – Genetic Genealogy