This is the third article in a series about mitochondrial DNA.
The first two articles are:
This third article focuses on haplogroups. They look so simple – a few letters and numbers – but haplogroups are a lot more sophisticated than they appear and are infinitely interesting!
What can you figure out about yours and what secrets will it reveal? Let’s find out!
What is a Haplogroup?
A haplogroup is a designation that you can think of as your genetic clan reaching far back in time.
My mitochondrial haplogroup is J1c2f, and I’ll be using this as an example throughout these articles.
The description of a haplogroup is the same for both Y and mitochondrial DNA, but the designations and processes of assigning haplogroups are different, so the balance of this article only refers to mitochondrial DNA haplogroups.
Where Did I Come From?
Every haplogroup has its own specific history.

Looking at my DNA Migration Map at Family Tree DNA, I can see the path that haplogroup J took out of Africa.

This map is interactive on your personal page, so you can view your or any other haplogroup highlighted on the map.

On the frequency tab of the Migration Map, you can view the frequency of your haplogroup in any specific location.

On my Mutations tab, I’m provided with this information:
The mitochondrial haplogroup J contains several sub-lineages. The original haplogroup J originated in the Near East approximately 50,000 years ago. Within Europe, sub-lineages of haplogroup J have distinct and interesting distributions. Haplogroup J1 is found distributed throughout Europe, from Britain to Iberia and along the Mediterranean coast. This widespread distribution strongly suggests that haplogroup J1 was part of the Neolithic spread of agriculture into Europe from the Near East beginning approximately 10,000 years ago.
Stepping-Stones back in Time
The haplogroup designation itself is a stepping-stone back in time.
Looking at my full haplogroup, J1c2f, we see 5 letters or numbers.
The first letter, J, is my base haplogroup, and each letter or digit after that will be another step forward in time from the “mother” haplogroup J.
Therefore, 1 is a major branch of haplogroup J, c is a smaller branch sprouting off of J1, 2 is a branch off of J1c, and f is the last leaf, at least for now.
Ages
In the supplementary data for the article, A “Copernican” Reassessment of the Human Mitochondrial DNA Tree from its Root, by Doron M Behar et al, published in the Journal of Human Genetics on April 6, 2012, he provides age estimates for the various haplogroups and subhaplogroups identified at that time.
My haplogroup breakdown is shown below.
|
Haplogroup |
Time Estimate (Years) | SD (standard deviation in years) |
| J | 34,258.3 | 4886.2 |
| J1 | 26,935.1 | 5272.9 |
| J1c | 13,072.3 | 1919.3 |
| J1c2 | 9762.5 | 2010.7 |
| J1c2f | 1926.7 |
3128.6 |
- Time estimate means how long ago this haplogroup was “born,” meaning when that haplogroup’s defining mutation(s) occurred.
- SD, standard deviation, can be read as the range on either side of the time estimate, with the time estimate being the “most likely.” Based on this, the effective range for the birth of haplogroup J is 29,372.1 – 39,144.5. In some of the most current haplogroups, like J1c2f, the lowest age range is a negative number, which obviously can’t happen. This sometimes occurs with statistical estimates.
The first question you’re going to ask is how can these age estimates be so precise? The answer is that these are statistical calculations – because we can’t travel back in time.
What Came Before J?
Clearly J is not Mitochondrial Eve, so what came before J?
In the paper announcing the latest version (Build 17) of the Phylotree by van Oven, meaning the haplotree for mitochondrial DNA, this pedigree style tree was drawn to show the backbone plus 25 subtrees.
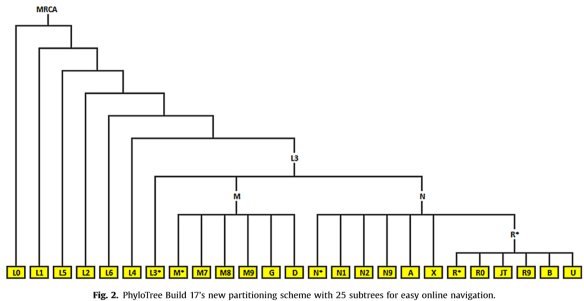
Haplogroup J descended from JT, fourth from right on the bottom right.
The MRCA, most recent common ancestor at the root of the tree would be the RSRS (Reconstructed Sapiens Reference Sequence), known colloquially as Mitochondrial Eve.
Branches and Names
Haplogroups were named in the order they were discovered, using the alphabet, A-Z (except O). Branches are indicated by subsequent numbers and letters. Build 17 of the phylogenetic tree includes 5437 branches, increasing from 4809 in build 16.
Occasionally branches are sawed off and reconnected elsewhere, which sometimes plays havoc with the logical naming structure because they are renamed completely on the new branch. This happened when haplogroup A4 was retired in Build 17 and is now repositioned on the tree as haplogroup A1. I wrote about this in the article, Family Tree DNA’s Mitochondrial Haplotree.
It’s easier to see the branching tree structure if you look at the public mitochondrial haplotree on the Family Tree DNA website. Scroll to the very bottom of the main Family Tree DNA page, here, and click on mtDNA haplotree.

You can search for your haplogroup name and track your ancestral haplogroups back in time.

J1c2f is shown below on the tree, with haplogroup J at the top.
Where in the World?
Whether you’ve tested at Family Tree DNA or not, you can view this tree and you can see the location of the earliest known ancestor of people who have tested, agreed to sharing and have been assigned to your haplogroup.
You can mouse over the little flag icons or click on the 3 dots to the right for a country report.
![]()
The country report details the distribution of the earliest known ancestors where people on that branch, and those with further subbranches are found.
J1c2f is the lowest leaf on this branch of the tree, for now, so there is no difference in the columns.
However, if we look at the country report for haplogroup J1c2, the immediate upstream haplogroup above J1c2f, you can see the differences in the columns showing people who are members of haplogroup J1c2 and also downstream branches.
I wrote more about how to use the new public tree here.
Haplogroup Assignment Process
There’s a LOT of confusion about haplogroup assignments, and how they are generated.
First, the official mitochondrial tree is the Phylotree, here. Assigning new haplogroups isn’t cut and dried, nor is it automated today. The Phylotree has been the defacto location for multiple entities to combine their information, uploading academic samples to GenBank, a repository utilized by Phylotree for all researchers to use in the classification efforts. You can read more about GenBank here. Prior to Phylotree, each interested entity was creating their own names and the result was chaotic confusion.
Individuals who test at Family Tree DNA can contribute their results, a process I’ll cover in a future article.
The major criteria for haplogroup assignments are:
- Three non-familial sequences that match exactly. Family mutations are considered “private mutations” at this time.
- Avoidance of regions that are likely to be unstable (such as 309, 315 and others,) preferably using coding region locations which are less likely to mutate.
- Evaluating whether transitions, transversions and reversions are irrelevant events to haplogroup assignment, or whether they are actually a new branch. I covered transitions, transversions and reversions here.
Periodically, the Phylotree is updated. The current version is Build 17, which I wrote about here.
The Good, the Bad and the Ugly
While change and scientific progress is a good thing, it also creates havoc for the vendors.
For each vendor to update your haplogroup, they have to redo their classification algorithm behind the scenes, of course, then rerun their entire customer database against the new criteria. That’s a huge undertaking.
In IT terms, haplogroups are calculated and stored one time for each person, not calculated every time you access your information. Therefore, to change that data, a recalculation program has to be run against millions of accounts, the information stored again and updating any other fields or graphics that require updating as a result. This is no trivial feat and is one reason why some vendors skip Phylotree builds.
When you’re looking at haplogroups at different vendors, it’s important to find the information on your pages there that identify which build they are using.
Vendors who only test a few locations in order to assign a base or partial haplogroup may find themselves in a pickle. For example, if a new Phylotree build is released that now specifies a mutation at a location that the vendor hasn’t tested, how can they upgrade to the new build version? They can’t, or at least not completely accurately.
This is why full sequence testing is critically important.
Haplogroup Defining Mutations

Using the Build 17 table published by Family Tree DNA that identifies the mutations required to assign an individual to a specific haplogroup or subhaplogroup, you can determine why you were assigned to a specific haplogroup and subgroups.
Mutations in Different Haplogroups are Not Equal
What you can’t do is to take mutations out of haplogroup context for matching.
Let’s say that someone in haplogroup H and haplogroup J both have a mutation at location G228A.

That does NOT mean these two people match each other genealogically. It means that the two different branches of the mitochondrial tree, haplogroup J and haplogroup H individually developed the same mutation, by chance, over time. In other words, parallel, disconnected mutations.
It may mean that both individuals simply happen to have the same personal mutations, or, it could mean that eventually these values could become haplogroup defining for a new branch in one or the other haplogroup.
How Common Are Parallel Mutations?
From the Build 17 paper again, this table shows us the top recurrent mutations after excluding insertions, deletions and location 16519. We see that 197 different branches of the tree have mutation T152C. My branch is one of those 197.

I think you can see, with location T152C being found in 197 different branches of the Pylotree why the only meaningful match between two people is within specific haplogroup subclades.
Within a haplogroup, this means that two people match on T152C PLUS all of the upstream haplogroup defining markers. Outside of a haplogroup, it’s just a chance parallel mutation in both lines.
Therefore, if another person in haplogroup J1c2f and I match a mutated value at the same location, that could be a very informative piece of genealogical information.
Partial and Full Haplogroups
Some vendors, such as 23andMe and LivingDNA provide customers with partial haplogroups as a part of their autosomal offering.
| Family Tree DNA (full haplogroup) | 23andMe | LivingDNA |
| J1c2f | J1c2 | J1c |
23andMe and LivingDNA provide partial haplogroups because they are not testing all of the 16,569 locations of the mitochondrial DNA. They are using scan technology on a chip that also processes autosomal DNA, so the haplogroup assignment is basically an “extra” for the consumer. Each chip location they use for mitochondrial (or Y) DNA testing for haplogroups is one less location that can be used for autosomal testing.
Therefore, these companies utilize what is known as target testing. In essence, they test for the main mutations that allow them to classify people into major haplogroups. For example, you can see that LivingDNA tests the mutations through the J1c level, but not to J1c2, and 23andMe tests to J1c2 but not J1c2f. If they tested further, my haplogroup designation would be J1c2f, not J1c or J1c2.
For full sequence testing, complete haplogroup designation and matching, I need to test at Family Tree DNA. They are the only vendor that provides the complete package.
Matching

Family Tree DNA provides matching of customer results. Consumers can purchase the mtPlus product, which tests only the HVR1/HVR2 portion of the mitochondria, or the mtFull product which tests the entire mitochondria. I recommend the mtFull.
In addition to haplogroup information, customers receive a list of people who match them on their mitochondrial sequence.
Matches with genealogical information allow customers to make discoveries such as this location information, provided by Lucille, above:

Lucille’s earliest known ancestor, according to her tree, is found just 12.6 km, or 7.8 miles from the tiny German village where my ancestor was found in the late 1600s.
Of course, matching isn’t provided in the 23andMe and LivingDNA databases, so we can’t tell who we do and don’t match genealogically, but haplogroups alone are not entirely useless and can provide great clues.
Haplogroups Alone
Haplogroups alone can be utilized to include or eliminate people for further scrutiny to identify descendancy on a particular line.

For example, at Family Tree DNA, I can utilize the advanced matching tool to determine whether I match anyone on both the Family Finder autosomal test AND on any of the mitochondrial DNA tests.
My match on both tests, Ms. Martha, above, has not tested at the full sequence level, so she won’t be shown as a match there. It’s possible that were she to upgrade that we would also match at the full sequence level. It’s also possible that we wouldn’t. Even an exact mitochondrial match doesn’t indicate THAT’s the line you’re related on autosomally, but it does not eliminate that line and may provide useful clues.
If my German match, Lucille and I had matched autosomally AND on the full sequence mitochondrial test, plus our ancestors lived 7 miles apart – those pieces of evidence would be huge clues about the autosomal match in addition to our mitochondrial match.
Alas, Lucille and I don’t match autosomally, but keep in mind that there are many generations between Lucille and me. If we had matched autosomally, it would have been a wonderful surprise, but we’d be expected not to match given that our common ancestor probably lived sometime in the 1600s or 1700s.
If I’m utilizing 23andMe and notice that someone’s haplogroup is not J1c2, the same as mine, then that precludes our common ancestral line from being our direct matrilineal line.
At GedMatch, people enter their haplogroup (or not) by hand, so they enter their haplogroup at the time they upload to GedMatch. It’s possible that their haplogroup assignment may have changed since that time, either because of a refined test or because of a Build number update. Be aware of the history of your haplogroup. In other words, if your haplogroup name changed (like A4 to A1), it’s possible that someone at GedMatch is utilizing the older name and might be a match to you on that line even though the haplogroup looks different. Know the history of your haplogroup.
Perhaps the best use of haplogroups alone is in conjunction with autosomal testing to eliminate candidates.
For example, looking at my match with Stacy at 23andMe, I see that her haplogroup is H1c, so I know that I can eliminate that specific line as our possible connection.

At Family Tree DNA, I can click on any Family Finder match’s profile to view their haplogroup or use the Advanced matching tool to see my combined Family Finder+mtDNA matches at once.

Haplogroups and Ethnicity
My favorite use of haplogroups is for their identification of the history of the ancestral line. Yes, in essence a line by line ethnicity test.
Using either your own personal results at Family Tree DNA, or their public haplotree, you can trace the history of your haplogroup. In essence, this is an ethnicity test for each specific line – and you don’t have to try to figure out which line your specific ancestry came from. It’s recorded in the mitochondrial DNA of each person. I’ve created a DNA pedigree chart to record all my ancestors Y and mitochondrial DNA haplogroups.
Ancestor DNA Pedigree Chart
Using Powerpoint, I created this DNA pedigree chart of my ancestors and their Y and mitochondrial DNA.

You can see my own mitochondrial DNA path to the right, in red circles, and my father’s Y DNA path at left, in blue boxes. In addition to Y DNA, all men have mitochondrial DNA inherited from their mother. So you can see my grandfather, William George Estes inherited his mitochondrial DNA from his mother Elizabeth Vannoy, who inherited it from Phoebe Crumley whose haplogroup is J1c2c.
This exercise disproved the rumor that Elizabeth Vannoy was Native American, at least on that line, based on her haplogroup. You can view known Native American haplogroups here.
So Elizabeth Vannoy and her mother, Phoebe Crumley, and I share a common ancestor back in J1c2 times, before the split of J1c2c and J1c2f from J1c2, so roughly 2,000 years ago, give or take a millennia.
Haplogroup Origins
My own haplogroup J is European. That’s where my earliest ancestor is found, and it’s also where the migration map shows that haplogroup J lived.

The information provided on my Haplogroup Origins page shows the location of my matches by haplogroup by location. I’m only showing my full sequence matches below.
Generally, the fewer locations tested, at the HVR1 or HVR1+HVR2 levels, the matches tend to be less specific, meaning that they may reach thousands of years back in time. On the other hand, some of those HVR1/HVR2 matches may be very relevant, but it’s unlikely that you’ll know unless you have a rare value in the HVR1/HVR2 region meaning few matches, or both people upgrade to the full sequence test.
You can see by the information above that most of my exact matches are distributed between Sweden and Norway, which is a very specific indicator of Scandinavian heritage ON THIS LINE alone.
By contacting and working with my matches of a genetic distance of 1, 2 and 3, I determined, based on the mutations, that the “root” of this group originated in Scandinavia and my branch traveled to Germany.
This is more specific than any ethnicity test would ever hope to be and reaches back to the mid-1600s. Better yet, I can make this same discovery for every line where I can find an individual to test – effectively rolling back the curtain of time.
Ancestral Origins
Haplogroup Origins can be augmented by the Ancestral Origins tab which provides you with the ancestral location of your matches’ most distant known ancestor.
Again, exact matches are going to be much more relevant to you, barring exceptions like heteroplasmies (covered here), than more distant matches.
New Haplogroup Discoveries
You might wonder, when looking at your results if there are opportunities for new haplogroup subgroups. In my case, there are a group of 33 individuals who match exactly and that include many common mutations in addition to the 11 locations in my results that are currently indicated as haplogroup identifying, indicated in red below.
My haplogroup defining mutation at A10398G! is a reversion, meaning that it has mutated back to the ancestral value, so we don’t see it above, because now it’s “normal” again. We just have to trust the ancestral branching tree to understand that upstream, this mutation occurred, then occurred a second time back to the normal or ancestral value.
The two extra mutations that everyone in this group has may be enough to qualify for a new haplogroup, call it “1” for purposes of discussion – so it could be named J1c2f1, hypothetically. However, there may be other sub-haplogroups between f and 1, so it’s not just a matter of tacking on a new leaf. It’s a matter of evaluating the entire tree structure with enough testers to find as many sub-branches as possible.
Attempting to assign or reassign branches based on a few tests and without a full examination of many tests in that particular branching haplotree structure would only guarantee a great deal of confusion as the new branch names would have to be constantly changed to accommodate new branching tree structures upstream.
This is exactly why I encourage people to upload their results to GenBank. I’ll step through that process in our last article.
What’s Next?
My next article in this series, in a couple weeks, will be Mitochondrial DNA: Part 4 – Techniques for Doubling Your Useful Matches. I more than doubled mine. There’s a lot more available than meets the eye at first glance if you’re willing to do a bit of digging.
But hey, we’re genealogists – and digging is what we live for!
______________________________________________________________
Disclosure
I receive a small contribution when you click on some (but not all) of the links to vendors in my articles. This does NOT increase the price you pay but helps me to keep the lights on and this informational blog free for everyone. Please click on the links in the articles or to the vendors below if you are purchasing products or DNA testing.
Thank you so much.
DNA Purchases and Free Transfers
- Family Tree DNA
- MyHeritage DNA only
- MyHeritage DNA plus Health
- MyHeritage FREE DNA file upload
- AncestryDNA
- 23andMe Ancestry
- 23andMe Ancestry Plus Health
- LivingDNA
Genealogy Services
Genealogy Research
- Legacy Tree Genealogists for genealogy research










Oh I have been just reading everything and Just got my early results HRV 1 and 2 they say V17 build 17 my daughter on 23 and me is just V. when I last looked a while back , Now I see why it will be different .,…
I took the mtFull Sequence 12/11/2018. I have NO matches? I’ve clicked every tab at FTDNA for my mt test and this is the result.
mtDNA – Results
Haplogroup – H-C16291T
Your Origin
mtDNA – Haplogroup Origins
HVR1 MATCHES
No Matches Found
HVR1 AND HVR2 MATCHES
No Matches Found
HVR1, HVR2, AND CODING REGION MATCHES
GENETIC DISTANCE -2
Haplogroup Country Comments Match Total
H-C16291T – 1
(there is nothing to click for this one. What do I do with it?)
mtDNA – Ancestral Origins
HVR1 MATCHES
No Matches Found
HVR1 AND HVR2 MATCHES
No Matches Found
HVR1, HVR2, AND CODING REGION MATCHES
No Matches Found
MATCHES MAPS
There are no locations available for your matches at this testing level. Please try a different testing level.
You must have several rare mutations. If you read the second article, you can use that to see.
You explanation of the less precise testing of mtDNA haplogroups at Living DNA and 23andMe and the resulting broader haplogroups makes sense. But my experience has been different.
I took the mtDNA full sequence test at FTDNA and I am haplogroup H13a1a1.
My mother tested at Living DNA and she has the same estimated haplogroup: H13a1a1
BUT we both tested and 23andMe and our estimated haplogroup there is longer:
H13a1a1a
Do you have any explanation of why this would be?
You can look at the version of the tree they are using and the locations they are using to calculate and compare to the latest version of the tree. It’s also possible there is a misread involved.
Thank you.
I love your colored Haplogroup tree. I am E-M35 and H2a2a1. I would like to create a similar tree, but I have no idea how you found the haplogroups for your distant ancestors.
You test living relatives. I have tested males for 5 of my Y-chromosome lines. I have done mtDNA tests for myself and two others so I know the mtDNA of 3 of my 4 grandparents. I am going to do a chart like Roberta’s.
Or check the projects to find people who have tested from your line.
This week I had weird Mt-DNA results I bothered to notice, thanks to your blogs.
I received a new HVR2 match at Family Tree DNA. I had been ignoring most of them, since I have a number of exact matches to look at. My son did a full Mt-DNA test (my Mt-DNA), but many other matches did not do full tests. I decided to look at this new match, and I almost fainted. I know the unusual surname given as that person’s rather recent ancestor. It is the maiden name of my husband’s uncle’s wife, now deceased. There is no tree attached, and a made-up user name. Maybe the tester was my husband’s female cousin, or cousin’s child, who I do not know? So I researched the uncle’s wife’s mother’s line back as far as possible from curiosity. Her father was from Germany, with other same surname matches unlikely, but the mother was not from Germany. That woman’s farthest back female that I found married 1798, Madison County, Kentucky. My farthest back female married 1795, Madison County, Kentucky. That is a very big coincidence!
So, I researched surnames somehow connected to this new research. I recognize some of them. In fact, I have met people with some of these surnames, or have them as somehow related in my tree. I then searched these surnames for matches on my mother’s sister’s Family Finder results, and I found a few weak matches. It is time to contact my husband’s female cousins from this uncle, since a couple of them have trees at Ancestry.com., and have done some DNA testing somewhere. Maybe one can be persuaded to do a full Mt-DNA test, or share other information? I might find another generation back on my mother’s line, or who knows what other information?
I just had to share this very strange results with people who care about Mt-DNA results. You never know what you will find.
Thank you for doing blogs on Mt-DNA!
a note- I have had very little luck contacting matching DNA testers for further information. No matter how polite or concise the request, people do not answer. Is there any way this can be improved?
I wish. People seldom answer on any platform.
Sometimes, they don’t use anymore the email they wrote there, sometimes, the person managing the email is no longer with us.
Use the email, name and any other info showing for this match and try to see if you can come up with something on internet. I found the obituary of one of my father’s Y-DNA match that way and with the info there, I could make his tree and find how we connected.
From the phylotree pages:
“The mutations 309.1C(C), 315.1C, AC indels at 515-522, A16182c, A16183c, 16193.1C(C) and C16519T/T16519C were not considered for phylogenetic reconstruction and are therefore excluded from the tree.”
My paternal grand-mother only has seven extra mutation, all are .1 to .4 A or C on positions 309, 315 and 522, so not a single useful mutation since… 9617.5 years ago, give or take up to 2327.5 years. Not too helpful with within a genealogically relevant time frame.
Yet, most of her exact match are descendants from her last known ancestress and many who aren’t are from places where you’d expect French Canadians to have been.
Among my own mt-DNA matches, some have an extra 522.2C, but there are given to me as exact match regardless (mitosearch was useful for these kinds of details).
I would guess there is a certain threshold from which the computer reporte the mismatch or not. Do you have more info about it?
These are not considered for the construction of the tree, but are reported in your results and matching, except for 309 and 315 which are reported but matches will not be excluded because of them.
So excited! I got my results back less than two weeks ago: so I studied your articles, Roberta, joined a project like you said to do, played around some, and then I decided to just pull a report, to see if one of my perfect matches (I’d gotten full sequence testing results) might match me on Family Finder (atDNA). I had learned it is rare, and even more rare that any shared ancestors in question would be the same from an atDNA and an mtDNA match, but if I lucked out and found one, maybe the shared ancestor would be more recent. So I clicked on the orange letters, “advanced matches” and asked for those who matched me on HVR1, HVR2, FMS and Family Finder. I clicked on “yes” for “show only people I match in all selected tests” on the second line, and for the pull-down for “show matches for” I chose “Entire Database” since my other choice brought up no results. (I’m not above just seeing what happens when I do certain things lol) I was so lucky: I got three testers! And one had attached a partial tree! So I did some genealogy, and built my match’s maternal line out a bit until I saw, lo and behold, the SISTER of my 2GGM! My big goal is to find and verify more remote female line ancestors, but I’ve gotten a good start. Thank you Roberta! Now…I might get you to help me on the paternal line thing: it hasn’t been as easy lol!
I’m so excited for you!!!
what’s the difference between a GD1 and an Exact Match? i realized after looking at my matches, i do have one Exact Match. i did contact her, and we exchanged surnames. none of them looked familiar because the surname of my furthest direct line maternal ancestor is unknown. found out that her direct maternal ancestor was from Ireland like mine was.
A GD 1 means that one of you has a mutation that the other doesn’t.
Pingback: Mitochondrial DNA: Part 2 – What Do Those Numbers Mean? | DNAeXplained – Genetic Genealogy
Pingback: Mitochondrial DNA: Part 4 – Techniques for Doubling Your Useful Matches | DNAeXplained – Genetic Genealogy
Pingback: Mitochondrial DNA: Part 5 – Joining Projects | DNAeXplained – Genetic Genealogy
Is H13a1a1 known for being common in Jewish populations?
Not that I know of. Take a look at your ancestral and haplogroup origins.
Hi… I just received my test results for the MTDNA full sequence. I wanted to ask as Im H3. Have known I was H3 for years as I had the 23andme test done. However I know the h3 has many subgroups, but my mtdna just simple says H3, no specific subgroup. Was my context wrong in thinking this would give me a more specific haplogroup? Or do I have to dig in order to find it? Still learning. Thanks
It could be that PhyloTree had not identified groups with your mutations below H3. Assuming you and others have mutations and match. We are working to do this through the Million Mito project.
G228A is also present in U5b1b1-T16192C!, though I’m the only sample that has it.
Pingback: 4 Kinds of DNA for Genetic Genealogy | DNAeXplained – Genetic Genealogy
As already pointed out by someone else, 23andMe can and occasionally does go beyond assigning a shorthand “haplogroup”.
The most aggravating instance is how they assign some people who really carry N1b1b1 the N1b2 lineage – which is on a totally separate branch from the source subclade N1b – while they would’ve settled for assigning them to N1b if you were totally correct.
To assign someone to N1b2 means “going all the way” just as Family Tree DNA would, since that’s the terminal SNP on one of the 2 branches that split off from N1b, just like the terminal SNP on the other branch.
When I retire, I will study your articles. In the meantime, wanted to let you know that my mother is J1C2.