If you are a Family Tree DNA customer, and in particular, a male or manage male kits, you’re familiar with the Big Y test.
The Big Y test scans the entire gold standard region of the Y chromosome, hunting for mutations, called SNPs, that define your haplogroup with great precision. This test also discovers SNPs never before found. Those newly discovered SNPs may someday become new haplogroup branches as well. The Big Y test is how the Y DNA phylotree has been expanded from a few hundred locations a few years ago to more than 78,000, and along with that comes our understanding of the migration patterns of our ancestors.
We’re still learning, every single day, so testing new people continues to be important.
The Big Y is the logical extension of STR testing (panels 37, 67 and 111), which focus on genealogical matches, closer in time, instead of haplogroup era matches. STR locations mutate more rapidly than SNPs, so the STR test is more useful for genealogists, or at least represent an entry point into Y DNA testing. SNPs generally reach further back in time, showing us where are ancestors were before STR test results kick in. More and more, those two tests have some time overlap as more SNPs are discovered.
If you want to read more, I wrote about this topic in the article, “Why the Big Y Test?”. Ignore the pricing information at the end of that article, as it’s out of date today.
Before we talk about the new format of the Big Y results, let’s take a step back and look at the multiple reasons why Family Tree DNA created a new Big Y experience.
The first reason is that the human reference genome changed.
What is the Human Reference Genome?
The Human Reference Genome is a genetic map against which everyone else is compared. In essence, it’s an attempt to give every location in our genome an address, and to have them all line up on streets where they belong on a nice big chromosome by chromosome grid.
That’s easier said than done. Let’s look at why and begin with a little history.
Hg refers to the human reference genome and 38 is the current version number, released in December of 2013.
The previous version was hg19, released in February of 2009.
This seems like a long time ago, but each version requires extensive resources to convert data from previous versions to the newer version. Different versions are not compatible with each other.
You can read more about this here, here, here and here, if you really want to dig in.
Hg19, the version that we’ve been using until now, was based only on 13 anonymous volunteers from Buffalo, New York. Hg38 uses far more samples and resequences previously sequenced results as well. We learned a lot between 2009 when the previous version, hg19, was released and 2013 when hg38 was released.
Keeping in mind that people are genetically far more alike than different, sequencing allows most of the human genome to be mapped when the genomes of those reference individuals are compared in layers, stacked on top of each other.
The resulting composite reference map, regardless of the version, isn’t a reflection of any one person, but a combination of all of those people against which the rest of us are compared.
Areas of high diversity, in this case, Y SNPs, may differ from each other. It’s those differences that matter to us as genealogists.
In order to find those differences, we must be able to line up the genomes of the various people tested, on top of each other, so that we can measure from the locations that are the same.
Here’s an example. All 4 people in this table above match exactly on locations 1-7, 9- 10 and 13-15.
Locations 8, 11 and 12 are areas that are more unstable, meaning that the people are not the same at that location, although they may not match each other, hence the different colored cells.
From this model, we know that we can align most people’s results on the green locations where everyone matches everyone else because we are all human.
The other locations may be the same or different, but they can’t be aligned reliably by relying on the map. You can read more about the complexity of this topic here and a good article, here.
A New Model
The challenge is that between 2009 and 2013, new locations were discovered in previously unmapped areas of the genome.
Think of genome locations as kids sitting in assigned seats side by side in a row.
Where do we put the newly discovered kids?
They have to crowd in someplace onto our existing map.
We have to add chairs between locations. The white rows below represent the newly discovered locations.
When we add chairs, the “addresses” of the kids currently sitting in chairs will change. In fact, the address of everyone on the street might change because everyone has shifted. Many of the actual kids will be the same, but some will be new, even though all of the kids will be referenced by new addresses.
This is a very simplified conceptual explanation of a complex process which isn’t simple at all. In addition to addressing, this process has to deal with DNA insertions, deletions, STR markers which are repeats of segments, palindromic mutations as well as pseudo-autosomal regions of the Y chromosome. Additionally, not all reads or calls are valid, for a number of reasons. Due to all these factors, after the realignment is complete, analysis has to follow.
Suffice it to say that converting from one version to the next requires the data to be reanalyzed with a new filter which requires a massive amount of computational power.
Then, the wheat has to be sorted from the chaff.
Discovery
The conversion to hg38 has been a boon for discovery, already. For example, Dr. Michael Sager, “Dr. Big Y” at Family Tree DNA has been busily working through the phylotree to see what the new alignment provides.
In November, he mentioned that he had discovered correct placement for a new haplogroup, high in the R1b tree, that joined together several subclades of U106.
In hg19, U106 had 9 subclades, all of which then branched downwards.
However, in hg38, utilizing the newly aligned genome, Michael can see that U106 has been reconfigured and looks like this instead.
Look at the difference!
- Two new haplogroups have been placed in their proper location in the tree; Z2265 and BY30097.
- A2150 has been repositioned.
- Because of the placement of A2150 and Z2265, U106 now only has two direct branches.
- S19589 has been moved beneath Z2265
- The remaining 7 peach colored haplogroups in the old tree are now subclades of BY30097.
You may not know or realize that this shuffle occurred, but it has and it’s an important scientific discovery that corrects earlier versions of the phylotree.
Congratulations Dr. Sager!
So, how does the conversion to hg38 affect customers directly?
The Conversion
In or about October 2017, Family Tree DNA began their conversion to hg38. Keep in mind that no other vendor has to do this, because no other vendor provides testing at this level for Y DNA, combined with matching.
Not only that, but there is no funding for their investment in resources to do the conversion. By that I mean that once you purchase the product, there is no annual subscription or anything else to fund development of this type.
Additionally, Family Tree DNA designed a new user interface for the enhanced Big Y which includes a new Big Y browser.
The initial conversion has been complete for some time, although tweaking is still occurring and some files are being reconverted when problems are discovered. Now, the backlog of tests that accumulated during the conversion and during the holiday sale are being processed.
So, what does this mean to the consumer? How do we work with the new results? What has changed and what does all of this mean?
It’s an exciting time. We’re all waiting for new matches.
I’m going to step through the features and functions one at a time, explaining the new functionality and then what is different, and why.
First Look
On your personal page, you have Big Y Results and Big Y Matches.
Either selection takes you the same page, but with a different tab highlighted.
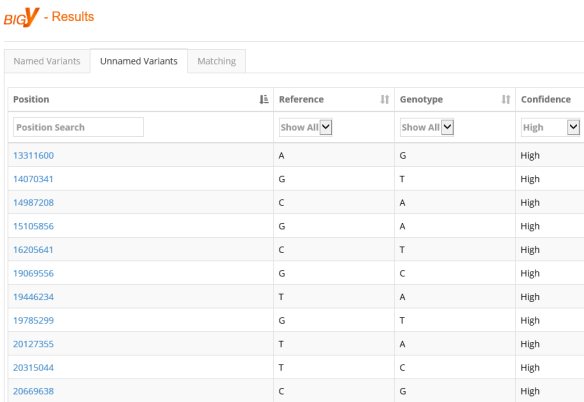
Named Variants
Named variants are SNPs that are already known and have been given SNP names.
At the bottom of the page, you can see that this person has 946 SNPs out of 77,722 currently on the tree. Many SNPs on the tree are equivalent to each other.
The information about each SNP on this page shows that it’s derived, meaning it’s a mutation and not ancestral which is the original state of the DNA.
If you look closely, you’ll see that some of the Reference and Genotype values are the same. You would logically expect them to be different. These are genuine mutations, but they are listed as the same because in hg19, the reference model, which is a composite, is skewed towards haplogroup R. In haplogroup R, these values are the same as the person tested (who is R-BY490), so while these are valid mutations on the tree of humanity, they are derived and found in all of haplogroup R. The same thing happens to some extent with all haplogroups because the reference sequence is a composite of all haplogroups.
The next column indicates whether the SNP has or hasn’t yet been placed on the Y tree.
The Reference column refers to the value at this address shown in the hg38 reference model, and the Genotype column shows the tester’s result at that location.
The confidence column shows the confidence level that Family Tree DNA has in this call. Let’s talk about confidence levels for a minute, and what they mean.
Confidence Levels
The Big Y test scans the Y chromosome, looking for specific blips at certain addresses. Every location has a “normal” blip for the Y chromosome as determined by the reference model. Any blips that vary from the reference model are flagged for further evaluation.
Blips can be caused by a mutation, a read error or a complex area of DNA, which is why there is a threshold for a minimum number of scans to find that same anomaly at any single location.
The area considered the “gold standard” portion of the Y chromosome which is useful genealogically is scanned between 55 and 80 times. Then the scans are aligned and compared to each other, with the blips at various locations being reported.
The relevance of blips can vary by location and what is known as density in various regions. In general, blips are not considered to be relevant unless they are recorded a minimum of 5 to 8 times, depending on the region of the Y chromosome. At that level, Family Tree DNA reports them as a medium confidence call. High confidence calls are reported a minimum of 10 times.
Some individuals and third-party companies read the BAM files and offer analysis, often project administrators within haplogroup projects. Depending on the circumstances, they may suggest that as few at 2 blips are enough to consider the blip a mutation and not a read error. Therefore, some third-party analysis will suggest additional haplogroups not reported by Family Tree DNA. Project administrators often collaborate with Dr. Sager to coordinate the placement of SNPs on the tree.
Therefore, at Family Tree DNA:
- You will see only medium and high confidence calls for SNPs.
- Over time, your Unnamed Variants will disappear as they are named and become Named Variants with SNP names.
- When Unnamed Variants become Named Variants, which are SNPs that have been named, they are eligible to be added to the Y tree.
- If the SNP added to the Y tree is below your present terminal SNP, you may one day discover that you have a new terminal SNP, meaning new haplogroup, listed on your main page. If the new SNP is within 5 upstream of your terminal SNP, looking backward up the tree, you’ll see it appear in your mini-tree on your personal page and on your larger Haplogroup and SNP page.
Unnamed Variants
Unnamed variants are newer mutations that have not yet been named as SNPs.
In order for a mutation to be considered a SNP, in true genetics terms, it has to be found in over 1% of the population. Otherwise, it’s considered a private, personal, family or clan mutation.
However, in reality, Family Tree DNA attempts to figure out which SNPs are being found often enough to warrant the assignment of a SNP number which means they can be placed on the haplotree of humanity, and which SNPs truly are going to be private “family mutations.” Today, nearly all mutations found in 3 or more individuals that are considered high confidence calls are named as SNPs.
Both named and unnamed variants are a good thing. New SNPs help expand and grow the tree. Personal or family SNPs can be utilized in the same fashion as STR markers. Eventually, as new SNPs are categorized and named, they will be moved from your Unnamed Variants page and added to your Named Variants page.
If you had results in the hg19 version, your unnamed variants will have changed. Just like those kids sitting on the bleachers, your old variants are either:
- Still here but with a new name
- Have been given SNP names and are now on your Named Variants list
The great news is that you’ll very probably have new variants too, resulting from the new hg38 reference model and more accurate alignment.
If you’re really a die-hard and want to know which hg19 locations are now hg38 locations, you can do the address conversion here. I am a die-hard but not this much of a die-hard, plus, I didn’t record the previous novel variant locations for my kits. Dr. Sager who has run this program tells me that you only need to pay attention to the two drop down menus specifying the “original” and “new” assemblies when utilizing this tool.
Y Chromosome Browser Tool
You’ve probably already noticed the really new cool browser tool, positioned tantalizingly to the right of both results tabs.
Go ahead and click on either a SNP name or an unnamed variant.
Either one will cause a pop up box to open displaying the location you’ve selected in the Big Y browser.
Utilizing the new Y chromosome browser tool, you can see the number of times that a specific SNP was called as positive or negative during the scan of your Y DNA at that specific location.
To see an example, click on any SNP on the list under the SNP Name column.
The Y chromosome browser tool opens up at the location of the SNP you selected.
The SNP you selected is displayed in pink with a downward arrow pointing to the position of the SNP. The other pink locations display other nearby SNP positions.
See that one single pink blip to the far right in the example above? That’s a good example of just one call, probably noise. You can see the difference between that one single call and high confidence reads, illustrated by the columns of pink SNP reads lined up in a row.
You can click on any of your SNP positions, named or unnamed, to see more information for that specific SNP.
Pink indicates that a mutation, or derived value, was found at that location as compared to the ancestral value found in the reference model.
Blue rows and green rows indicate that the forward (blue) or reverse (green) strand was being read.
The intensity of the colors indicates the relative strength of the read confidence, where the most intense is the highest confidence.
The value listed at the top, T, A, C or G is the abbreviation for the ancestral reference nucleobase value found in the reference population at that genetic location, and the value highlighted in pink is the derived (mutated) value that you carry.
Confidence is a statistical value calculated based upon the number of scans, the relative quality of that part of the Y chromosome and the number of times that derived value was found during scanning.
I love this new tool.
I hope that in the next version, Family Tree DNA will include the ability to look at additional locations not on the list.
For example, I was recently working on a Personalized DNA Report where the SNP below the tester’s terminal SNP was not called one way or another, positive or negative. I would have liked to view his results for that SNP location to see if he has any blips, or if the location read at all.
Matching
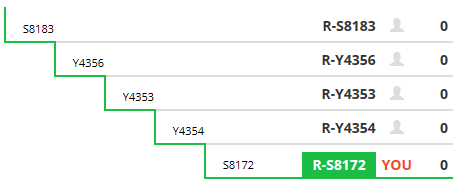
The third tab displays your Big Y matches and a mini-tree of your 5 SNPs at the end of your own personal branch of the haplotree.
Your terminal SNP determines the terminal (final or lowest) subbranch (on the Y-DNA haplotree) to which you belong.
On your mini-tree, your terminal SNP (R-BY490 above) is labeled YOU.
The number of people you match on those SNPs utilizing the new matching algorithm is displayed at each branch of the tree.
The matches shown above are the matches for this person’s terminal SNP. To see the people matching on the next branch above the terminal SNP, click on R-BY482.
The number listed beside these SNPs on your 5 step mini-tree is NOT the total number of people you match on that branch, only the number you match on that branch AFTER the matching algorithm is applied.
I put this in bold red, because based on the previous matching algorithm that managed to include everyone on your terminal SNP, it’s easy to presume the new version shows everyone in the system who matches you on that SNP – and it doesn’t necessarily. If assume it does or expect that it will, you’re likely to be wrong. There is a significant amount of confusion surrounding this topic in the community.
New Matching Algorithm
The Family Tree DNA matching algorithm has changed substantially. It needed to be updated, as the old matching algorithm had been outgrown with the dramatic new number of SNPs discovered and placed on the phylotree. Family Tree DNA created the original matching software when the Big Y was new and it was time for a refresh. In essence, the Big Y testing and tree-building has been successful beyond anyone’s wildest dreams and the matching routine became a victim of its own success.
Previously, Family Tree DNA used a static list of somewhere around 6,000 SNPs as compared to over 350,000 today, of which more than 78,000 have been placed on the tree. By the way, this SNP number grows with every batch of Big Y results because new SNPs are always found.
The previous threshold for mismatches was 4 SNPs. As time went on, this combination of a growing tree and a static SNP list caused increasingly irrelevant matches.
For example, in some instances, haplogroup U106 people matched haplogroup P312 people, two main branches of the R1b haplotree, because when compared to the old SNP list, they had less than 4 SNP mismatches.
The new Big Y matching routine expands as the new tree grows, and isn’t limited. This means that people who were shown as matches to haplogroups far upstream (e.g. P312/U106), whose common ancestor lived many thousands of years ago, won’t be shown as matches at that level anymore.
Many people had hundreds of matches and complained that they were being shown matches so distant in time that the information was useless to them.
The previous Big Y version match criteria was:
- 4 or less differences in Known SNPs (now Named Variants.)
- In addition, you could have unlimited differences in Unnamed Variants, then called Novel Variants.
Family Tree DNA has attempted to make the matching algorithm more genealogically relevant by applying a different type of threshold to matching.
In the current Big Y version, a person is considered a match to you if they have BOTH of the following:
- 30 or fewer differences in total SNPs (named and unnamed variants combined.)
- Their haplogroup is downstream from your terminal SNP haplogroup or downstream from your four closest parent haplogroups, meaning any of the 5 haplogroups shown on your 5 step mini-tree.
Here’s the logic behind the new matching algorithm threshold.
SNP mutations happen on the average of one every 100 years. This number is still discussed and debated, but this estimate is as good as any.
If your common ancestor through two men had two sons, 1500 years ago, and each line incurred 1 mutation every hundred years, at the end of 1500 years, the number of mutations between the two men would be approximately 30.
Family Tree DNA felt that 1500 years was a reasonable cutoff for a genealogical timeframe, hence the new matching threshold of 30 mutations difference.
The new match criteria is designed to reflect your matches that are most closely related to you. In other words, the people on your match list should be related to you within the last approximate 1500 years, and people not on your match list who have taken the Big Y are separated from you by at least 30 mutations.
There may be people in the data base that match you on your terminal SNP and any or all of the SNPs shown on your mini-tree, but if you and they are separated by more than 30 differences (including both named and unnamed variants) on the Y chromosome, they will not be shown as a match.
By clicking on the SNP name on your mini-tree, at right, you can see all of the people who match you with less than 30 differences total at each level, and who carry that particular Named Variant (SNP). The example shown above show this person’s matches on their terminal SNP. If they were to click on BY482, the next step up, they would then see everyone on their match list who is positive for that SNP.
On your match page, you can search for a specific surname, nonmatching variants or match date.
The Shared Variants column is the total number of shared variants you have with the match in question. According to the lab at Family Tree DNA, this number very high because it is reflective of many ancient variants.
You can also download your data from this page into a spreadsheet.
The Biggest Differences
What you don’t receive today, that you did receive before, is a comprehensive list of who you match on your terminal and upstream SNPs.
For example, I was working with someone’s results this week. They had no matches, as shown below.
However, when I went to the relevant haplogroup project page, I discovered that indeed, there are at least 4 additional individuals who do share the same terminal SNP, but the tester would never know that from their Big Y results alone, if they didn’t check the project results page.
Of course, it’s unlikely that every person who takes the Big Y test joins a Y DNA project, or the same Y DNA project. Even though projects will show some matches, assuming that the administrator has the project grouped in this manner, there is no guarantee you are seeing all of your terminal SNP matches.
Project administrators, who have been instrumental in building the tree can also no longer see who matches on terminal SNPs, at least not if they are separated by more than 30 mutations. This hampers their ability to build the Y tree.
This matching change makes it critical that people join projects AND make their results viewable to project members as well as publicly. Most people don’t realize that the default when joining projects is that ONLY project members can see their results in the project. In other words, the results are available in the public project, like the screenshot above.
You can read more about Family Tree DNA’s privacy settings here.
Another result of the matching algorithm change is that in some cases, one man may match a second man, but the second man does not show up on the first man’s match list.
I know that sounds bizarre, but in the Estes project, we have that exact scenario.
The chart above shows that none of the Estes Big Y participants match kit number 166011, also an Estes male, but kit 166011 does show matches to all of those Estes men.
Kit 166011 is the one to the far right on the pedigree chart above, and he is descended from a different son of Robert born in 1555 than the rest of the men. Counting from kit 166011 to Robert born in 1555 is 12 generations. Counting from kits 244708 and 199378 to Robert is 10 generations, so a total of 22 generations between those men.
Kits 366707, 9993 and 13805 are 11 generations from the common ancestor, so a total of 23 generations. Not only are these genealogically relevant, they carry the same surname.
The average of 30 mutations reaching to 1500 years doesn’t work in this case. The cutoff was about 1555, or 462 years, not 1500 years – so the matching algorithm failed at 30% of the estimated time it was supposed to cover. I guess this just goes to prove that mutations really don’t happen on any type of a reliable schedule – and the average doesn’t always pertain to individual family circumstances.
If you’re wondering if these men match on STR markers, they do.
In this case, the Big Y doesn’t show matches in a timeframe that STR markers do – the exact opposite of what we would expect.
One of the benefits of the Big Y, previously, was the ability to view people of other surnames who matched your SNP results. This ability to peer back into time informed us of where our ancestors may have been prior to where we found them. While this isn’t genealogy, per se, it’s certainly family history.
A good case in point is the Scottish clans and how men with different surnames may be related.
As a family historian I want to know who I match on my terminal SNP and the direct upstream SNPs so I can walk this line back in time.
What’s Coming
At the conference in Houston in November, Elliott Greenspan discussed a new direction for the Big Y in 2018. The new feature that all Big Y testers are looking forward to is the addition of STRs beyond the 111 marker panels, extracted from the Big Y as a standard product offering. Meaning free for Big Y testers.
The 111 and lower panels will continue to be tested on their current Sanger platform. Analysis of more than 3700 samples in the data base that have both the Big Y and 111 markers indicate that only 72 of the 111 STR markers can be reliably and consistently extracted from the Big Y NGS scan data. The last thing we want is unreliable NGS data being compared to our Sanger sequenced STR values. We need to be able to depend on those results as always being reliable and comparable to each other. Therefore, only STR markers above 111 will be extracted from the Big Y and the original 111 STR markers will continue to be sold in panels, the same as today.
However, because of the nature of scanning DNA as opposed to directly testing locations, all of the markers above 111 will not be available for everyone. Some marker locations will fail to read, or fail to read reliably. These won’t necessarily be the same markers, but read failure will apply to some markers in just about every individual’s scan. Therefore, these additional STR markers will be supplemental to the regular 111 STR markers. You get what you get.
How many additional markers will be available through Big Y? That hasn’t been finalized yet.
Elliott said that in order to reliably obtain 289 additional markers, they need to attempt to call 315. To get 489, they have to attempt more than 600, and many are less useful.
Therefore, speculating, I’d guess that we’ll see someplace between 289 and 489, the numbers Elliott mentioned.
Are you salivating yet?
Given that the webpage and display tools have to be redesigned for both individuals’ results, project pages and project administrators’ tools, I’d guess that we won’t see this addition until after they get the kinks worked out of the hg38 conversion and analysis.
It’s nice to know that it’s on the way though. Something to look forward to later in 2018.
In Summary
I know that the upgrade to hg38 had to be done, but I hated to see it. These things never go smoothly, no matter who you are and this was a massive undertaking.
I’m glad that Family Tree DNA is taking this opportunity to innovate and provide the community with the nifty new Y DNA browser.
I’m also grateful that they listen to their customers and make an effort to implement changes to help us along the genealogy path.
However, sometimes things fall into the well of unintended consequences. I think that’s what’s happening with the new matching routine. I know that they are continuing to work to tweek the knobs and refine the results, so you’re likely to see changes over the next few months. It’s not like there was a pattern or recipe anyplace. This has never been done before.
Here’s a list of changes and updates I’d suggest to improve the new hg38 Big Y experience:
- In addition to threshold matching, an option for direct SNP tree matching through the 5 SNPs shown on the participant’s 5 step mini-tree, purely based on haplotree matching. This second option would replace the functionality lost with the 30-mutation threshold matching today.
- A matches map of the most distant ancestors at each level of matching for both threshold matching and SNP tree matching.
- An icon indicating whether a Big Y match is an STR match and which level of STR panel testing the match has completed. This means that we could tell at a glance that a Big Y match has tested to 111 markers, but is only a match at 12.
- An icon indicating if the Big Y match has also taken the Family Finder test, and if they are a match.
- An icon on STR matches pages indicating that a match has taken a Big Y test and if they are a match.
- Ability to query through the Big Y browser to SNP locations not on the list of named or unnamed variants.
- Age estimates for haplogroups.
If you are seeing Big Y results that you find unusual or confusing, please notify Family Tree DNA support. There is a contact link with a form at the bottom of your personal page. Family Tree DNA needs to be aware of problems and also of customer’s desires.
Family Tree DNA has indicated that they are soliciting customer feedback on the new Big Y matching and tools.
Please also join a relevant haplogroup project as well as a surname project, if you haven’t already. Here’s an article, What Project Do I Join?, to help you find relevant projects.
If you think you have an unnamed variant that should be named and placed on the phylotree, your haplogroup project administrator is the person who will work with you to verify that the unnamed variant is a good candidate and submit the unnamed variant to Family Tree DNA for naming.
If you are a project administrator having issues, questions or concerns, you can contact the group projects team at groups@ftdna.com. Be sure that this address is in the “to” field, not the “cc” field as the e-mail will bounce otherwise.
Don’t forget that you can reference the Family Tree DNA Learning Center about your Big Y results.
Thank you to Michael Sager for his assistance with this article.
- ______________________________________________________________
Disclosure
I receive a small contribution when you click on some of the links to vendors in my articles. This does NOT increase the price you pay but helps me to keep the lights on and this informational blog free for everyone. Please click on the links in the articles or to the vendors below if you are purchasing products or DNA testing.
Thank you so much.
DNA Purchases and Free Transfers
- Family Tree DNA
- MyHeritage DNA only
- MyHeritage DNA plus Health
- MyHeritage FREE DNA file upload
- AncestryDNA
- 23andMe Ancestry
- 23andMe Ancestry Plus Health
- LivingDNA
Genealogy Services
Genealogy Research
- Legacy Tree Genealogists for genealogy research





















Roberta, I just wanted you to know that I did the Helix holiday offer electing for both Metabolism and Neanderthal(altho I had 23andme reading of latter). Insitome sent me 2 e-mails saying the results are ready, yet I cannot access this info until I give them a photo of me to “complete my profile” This was not disclosed to me when I sent sent my money and DNA sample. You might want to warm your readers since you gave them such a fulsome report.
I will forward this to them. I would presume that any photo would do.
Hi Carol – This is Neeraj Rao, co-founder and COO at Insitome. We are actively working to simplify the account setup process including adding a button to skip the optional profile information. For now, as Roberta pointed out, any image can be used to complete the photo upload step. There’s no requirement that it be an actual picture of you. We are happy to provide an image as well if needed.
We understand your concerns about this and take your feedback seriously. If you, or anyone else reading this, have questions about Insitome’s products or policies, please contact us at help@insito.me. Thank you!
The Privacy and Sharing section of this blog confused me. I had assumed that I had my settings correct in order for the project administrators to view my data. now, I’m not so sure. Given this change to the Big Y, what is the recommended setting for “Who can view my DNA results in group projects?” Thank you.
Project admins can view your Y results if you are in the project. The default is for other project members to be able to view your results too, BUT not the public. When I wrote this article, I referenced the public website of the project, and found 4 people there with matching SNPs. However, because the person’s DNA I was utilizing is in a project, I know there were actually more whose results are not set to be able to be viewed publicly.
My goodness, I can now actually look at my brother’s BigY results and understand the information that is there. Roberta, you have no idea how much you are appreciated and even then – that’s not enough. FTDNA should pay you and publish your information on their site.
Actually, I just sent them a copy and gave them permission to use it:) I don’t know if they will or not, but I do know that support often sends people to various articles. Different people learn in different ways. And thank you!
You’re welcome – sending FTDNA a copy is just another indication of your generosity. Okay, I won’t get carried away (I actually feel weepy after a year of trying to understand this stuff). Just thank you.
Thank you. Thank you. Thank you! Previously, I had nearly given up in despair when I saw my son’s list of 0s on his mini-matches page. Your explanation of the processes involved is so helpful in understanding the results and reminded me that the science of genetic genealogy is a work-in-progress and not the final word.
“science of genetic genealogy is a work-in-progress and not the final word” – best quote EVER!!!!
I suspect I must be missing something and will sound really stupid as a result — but here it goes. In your chart called “Named Variants” — “Big Y Results” — the top 4 SNPs a(nd some others below them) show that the “Reference” and the “Genotype” have the identical value. I thought that if you have the same value as the reference at a particular location, that you do not have the SNP that has been detected in others at that location. What am I missing?
Good question. I checked with Dr. Sager on this one. Your understanding is accurate, but in some cases, the mutation is real, but because the reference genome was derived (meaning it too had the mutation), the reference genome and derived value match. This happens often in haplogroup R because that was the reference genome for hg19. The example I used is haplogroup R.
I’m having a hard time grasping how anyone would know that the Reference has a derived value instead of the ancestral value. And if in fact they somehow knew that to be true — why wouldn’t the Reference be modified to have the ancestral value instead of a derived value. I had a similar situation with my Big Y testing using the old standard. When they looked into it, they said that it was a mistake and eliminated those “non-SNP” SNPs. I strongly suspect the above chart is also mistaken and Dr. Sager was simply too busy to investigate it fully.
If you think about this, if the reference panel is a composite, how to you account for the fact that the haplogroup R and the haplogroup A and the haplogroup E values are all different at a specific location? There can only be one “reference model.” We have this same issue with the CRS model in mtDNA. The CRS reference person was a subclade of H2, so while 16519C is a “mutation,” it’s not really – it’s that the reference model carried a mutation, meaning that the mutation at 16519 is actually the most common “mutation” in the world, because most people have it, because the reference model carried the mutation which is now compared to as “normal.”
I know nothing about Mt DNA so I won’t comment on that. But as to your other question, if Hg’s R, E and A have different values at a given location, I think there are 2 possible routes forward: 1) Which value do most Hgs agree with. Wouldn’t that likely be ancestral? If there were no clear winner from that analysis — wouldn’t the average of the earlier Hgs likely be ancestral? In other words, since we know DNA is reasonably stable — wouldn’t we assume the smallest number of mutations to get us from ancestral to present?
The point is that the reference model is determined outside of FTDNA’s venue. Therefore, all they can do is compare to the model and note the mutations as known to exist.
If I want my cousin to take a test for his Y, should it be Family Tree?
Yes, the link is on the sidebar. They are the only ones that do testing and matching.
Wow – thanks for explaining all of these changes – I will probably be re-reading this article several times over the coming weeks as I struggle to figure out all of these ins and outs and ups and downs and sideways connections!
In my Portuguese Big Y kits, I am also seeing far more mutations than the average and so this matching algorithm will not be so welcome until we have far better coverage of the population. I think YFull’s analysis of the BAM files will be ever more important to understand who you match and when.
An addition to the wish list would be separating “Ancestral Surname” matches from “Cousin Surname” matches, to catch our ancestors’ female descendents’ married surnames.
The Y chromosome does not descend through the female line – only the direct paternal line straight back in time, so this division isn’t possible or necessary. Unless I’m misunderstanding your request.
“If you’re really a die-hard and want to know which hg19 locations are now hg38 locations”
for the named SNP’s Go to http://www.snpdata.com
And choose:
Compare Snp Data Hg19 And Hg38 positions
This is for BigY, what about Fullgenome?
Do you mean what about working with the full genome using hg38?
Thanks for the review, it will be handy when I’ll join the fun in a few weeks.
Crossing my fingers that I will finally have matches (I mean other than the Y-12 bunches).
Thank you for your detailed walk through of the updated Big Y features, Roberta. I appreciate your clear step-by-step explanation.
Has FTDNA said anything more specific about the likelihood that Big Y would provide age estimates for haplogroups in 2018? I’m most interested in this capability and wondered about a timeline and what’s involved in setting that up with an ever-evolving Y tree.
No, I haven’t heard anything. It’s just on my wish-list:)
Hello Roberta,
Thank you for the informative article. From what I’ve seen on the various forums I visit, a lot of people have been complaining that the new matching threshold is too stringent and it’s blocking people from seeing their distant & ancient matches. If I’m understanding you correctly, your first suggestion would remedy that situation. Besides, sending them your article, have you spoken to anyone at FT about the improvements you have in mind?
“In addition to threshold matching, an option for direct SNP tree matching through the 5 SNPs shown on the participant’s 5 step mini-tree, purely based on haplotree matching. This second option would replace the functionality lost with the 30-mutation threshold matching today.”
Yes, they have a copy and I am in communication with them. I would encourage others as well.
Thank you Roberta, I have and I’ve been encouraging others to do so.
Hi Roberta,
Many thanks for explaining why sometimes a Variant has the same Reference and Genotype values. It’s an important point and one that left me scratching my head for ages. This is the first time I’ve seen a good explanation of this point.
hi
In regards to matching with others,
I originally had 3 matches under ftdna hg19 and now have zero matches with hg38, but yfull has me with 6 matches ( both hg19 and hg38 ) of which 5 matches are identical with ftdna project group.
which one is accurate now in matches ?
The matching is different. FTDNA is showing you genealogically relevant matches within about 1500 years and YFull is showing all matches on a particular SNP.
Roberta, how many unnamed variants are there for each of the Estes kits 9993, 13805, 166011, 199378, 244708, 366708, and N167192?
Has Dr. Michael Sager explained why none of the Estes Big Y participants match kit number 166011, also an Estes male, but kit 166011 does show matches to all of those Estes men? It looks like a glitch to me since the number of non-matching variants has to be the same between two people regardless of which of the two kits is looked at.
Have the new VCF files from the Estes kits from FTDNA been sent to the Y-DNA Data Warehouse so that Alex Williamson can analyze them and update the info at http://www.ytree.net/DisplayTree.php?blockID=550 ? It would be interesting to see a comparison of unnamed variants at FTDNA and the number of variants that show for each of those kits at Ytree. For example, kit 166011 currently has 6 variants at http://www.ytree.net/SNPinfoForPerson.php?personID=800
I am working with Dr. Sager on this. I will hopefully be publishing something soon as a follow up.
This is a great explanation Roberta, thank you!
After reading your post, I was wondering how the 111 STR matches and the BigY SNP matches work together. For example (within a surname group), one man (FM), who has a completely documented lineage back to the late 1100s, is both a BigY match and a 102/111 STR match to DM. FM and DM appear to have branched off from one another on the Big Tree around 1200 AD (descent from two brothers). Another man, BM, (who is from FM’s general line) is a 104/111 match to DM. How do you make sense of the relationship between an individual’s BigY SNP matches and their STR 111 matches? I know that some of the difficulty is that not all the men in this surname group have done the BigY, but 10 out of DM’s 13 BigY matches are not in his 111 STR matches, but 3 of the men are and none of them are on DM’s branch that split off around 1200 AD. I’ve been searching your blog and the internet in general and can’t find any references or teaching materials on this so that I might better understand how to make sense of it. Thanks so much!
Part of the reason nothing exists is because this is still so new and there aren’t any hard and fast rules. Men will match on SNPs much further back in time than they will match on STR markers. In some cases, with younger SNPs, you will see some anomalies on this, but not often.
Thanks Roberta! The SNP in question is a younger SNP, dated about 1200 AD. It’s interesting to see SNPs and STRs beginning to converge.
Those are the problem ones.
An interesting talk on line (which seems to have disappeared) by James Irvine at the just concluded Back to our Past meeting in Belfast (Genetic Genealogy Ireland group on Facebook) posits that the Big Y is a waste of money and that the individual STP packs are better. I tend to agree with you and did get the Big Y (which just arrived) But I wonder if I did right.
It all depends on what you want to do. If you want to confirm specific SNPs, ONLY, then SNP packs are fine. But you will never make a new discovery with a SNP pack. If you are interested in seeing if your DNA contains any newly discovered mutations, then a SNP pack won’t do that for you and the Big Y is the way to go. Look at it another way – if people hadn’t taken the Big Y, there wouldn’t be any SNP packs because those SNPs still wouldn’t be discovered. It’s a matter of personal preference.
Thanks, Roberta. I didn’t explain that James Irvine is the moderator of Irvine Project at FTDNA and really gave a fine presentation, overall, on the methodology of entwining DNA with family trees. He also said Y-37 was sufficient and that going up to 67 & 111 was marginal at best. (A true son of Scotland). Later he allowed there was some usefulness in them. He’s shown good results within his project so I’m not criticizing him, but I feel happy having gone to Y-111 & Big Y. Besides, it’s just the grandkids inheritance, anyway…
Thanks
Question – I did my BigY in 2016 and now I see …results completed 10/20/2017…….but my Bam file is still dated 2016………….have I been upgraded from hg19 to hg38 ?
regards
The gh38 BAM file shave not been released yet.
Note: I also have no confidence column in any report
Pingback: Whole Genome Sequencing – Is It Ready for Prime Time? | DNAeXplained – Genetic Genealogy
What is the age of S19589 group?
Pingback: Exciting New Y DNA Haplogroup D Discoveries! | DNAeXplained – Genetic Genealogy
Pingback: Family Tree DNA Names 100,000 New Y DNA SNPs | DNAeXplained – Genetic Genealogy
Thanks for an informative article. I appreciate you helping us learn this stuff. My question is about the comment: “SNP mutations happen on the average of one every 100 years.” Does this mean a new haplogroup marker would be created with each such mutation?
Yes, on average.
Roberta,
Thank you for your explanation. But I have a question on Non-Matching Variants. What are they not matching to? I.e. – I am comparing James and Ernest of the same surname. When I look at the Non-Matching Variants from James’ side, then it lists 13 mutations under Ernest. If I view from Ernest’s side, it lists the identical 13 mutations for Jame. I assume if they are non-matching, then they should be different. Or is this comparing to some other reference? Thanks
These are the variants where they don’t match each other.
I have one match in my Blocktree (Y-67 GD 7, Big Y 0/386) who has no (zero) non-maching variants. On the other hand he does not come up on my matches when searching by surname in Big Y matches. Why?
I don’t know. I would suggest contacting support.
My Big Y results have arrived today. I’m I-S14393. This article is very informative and detailed, thanks!