I’m so pleased to introduce The Million Mito Project and to explain why you should join in the quest to trace the family tree of womankind. The Million Mito Project depends upon you!

I was honored to announce The Million Mito Project at RootsTech on February 29, 2020. I’m sharing my slides with you here, along with the narrative.

The Million Mito Project is a collaborative citizen science project to update the phylotree of womankind.
Team members include:
- Miguel Vilar, Lead Scientist of the National Geographic Society’s Genographic Project
- Paul Maier, Population Geneticist at FamilyTreeDNA
- Goran Runfeldt, Head of Research and Development at FamilyTreeDNA
- Roberta Estes, DNAeXplain, scientist, genetic genealogist, Genographic Project Affiliate Researcher
Genographic Project Public Participation Phase Ended
The Genographic Project was launched on April 13, 2005 as a 5-year non-profit citizen-science-fueled research project with testing performed by the lab at Family Tree DNA. Unimaginably successful, the Genographic Project celebrates its 15th anniversary in 2020. I can’t tell you what a wonderful opportunity it has been to be involved with the Genographic project from its inception, and to be a part of the next chapter in this legacy of humanity.
It’s important to note that the public participation phase of the Genographic Project came to an end in 2019, meaning that kits can no longer be purchased. The Genographic database will remain online through June 30, 2020, but will be shuttered down after that time.
If you do not retrieve your Genographic results, or transfer them elsewhere before June 30, 2020, they will no longer be available to you. You can read more, here.
Even though the public participation phase has come to an end, the scientific study continues. That’s the legacy of the Genographic Project, the gift that keeps on giving.
Why DNA?
Every human alive carries the mitochondrial DNA inherited from their direct matrilineal line – your mother, her mother, her mother on up the tree into the mists of history.
This means that the Million Mito Project is relevant for every human living today – and carries critical genealogical and historical information passed to us from our matrilineal ancestors. Mitochondrial DNA is the one way that every human alive can see beyond the confines of records and available genealogy by using the gift of DNA that our ancestors bestowed up us.
It was 67 years ago on February 28th, the day before my RootsTech presentation, that Watson and Crick “discovered” DNA at Cambridge University in the Cavendish Lab.
“Discovered” is in quotes because there remains significant controversy about the fact that their discovery was predicated upon the research of Rosalind Franklin, who died and never received proper credit for her co-discovery. So really, the discovery should be credited to Watson, Crick and Franklin.

The slide above shows me standing in the doorway of the building in which this revolutionary discovery was made a few years before I was born. Ironically, it was DNA that drew me to England.
Y DNAwas the tool that allowed the US Speaks family to connect with the UK Speaks family in Lancashire through a man who had immigrated to New Zealand. Without Y DNA, the relevant deeply-buried records would not have been found, nor the genetic “glue” to tie records to people on three continents around the world, reuniting our widely-scattered family back in our ancestral homeland. Therein lies the amazing power of DNA.
Given my career choice, I absolutely had to visit the Cavendish Lab at Cambridge, a genetic “Mecca” of sorts, as well as the British Science Museum to see the infamous DNA model. This is where the DNA journey for all genetic genealogists began.
After their momentous discovery back in 1953, Watson and Crick walked the short distance to the Eagle Pub where they lunched regularly, shown above, at right, and excitedly announced to everyone within earshot that they had just discovered “the secret of life.”
Of course, 67 years ago, in the Eagle Pub, no one understood or cared.
We care, a lot, today. Beginning 20 years ago with the founding of FamilyTreeDNA, DNA fundamentally changed genealogy forever, allowing us to unravel mysteries that could never have been solved before.
Inseparable Technologies

Today, genealogy, genetic genealogy which focuses on the DNA aspects of genealogy and science are all inseparably intertwined. Scientific discoveries feed the genealogy and tests taken by genealogists fuel the science. It’s an infinite loop of discovery, education and unraveling.
Many times, the pieces of genealogical information we so desperately seek simply aren’t available in existing records, but we can piece together relationships and clues using the three different kinds of DNA: mitochondrial, Y DNA and autosomal. Each type of DNA has specific characteristics and provides us with unique information not obtainable any other way.
3 Kinds of DNA Address 3 Unique Challenges

Y DNA, inheritance path shown by the blue arrow above, is passed from father to son and therefore tracks back to a male’s direct patrilineal ancestors in their tree. The Y chromosome is only contributed to male children, who pass it on to their male children, not mixed with any DNA from the mother. Therefore, except for occasional mutations, Y DNA is identical from generation to generation.
The occasional mutations are what make it possible for us to use Y DNA and mitochondrial DNAas breadcrumbs, following them infinitely back into time, before records, and eventually, before written history by connecting those mutation breadcrumbs as dots.
Today, men test their Y DNA which follows the direct patrilineal line, which is the same as the surname line in western culture, although naming practices vary in different countries and parts of the world across time. Regardless, the Y DNAconnects male testers with their ancestors through their father’s, father’s, fathers’ line – in close relationships, meaning fathers and grandfathers, as well as distantly, into the history of clans and then before the advent of surnames.
In western cultures, men taking Y DNA tests expect at least some of their matches to carry the same or similar surnames, assuming other men from that line have also tested.
Only males can test for Y DNA, because only males carry a Y chromosome. Women need to ask their brothers, father, grandfather, uncles, etc. that represent the line they wish to test.
Mitochondrial DNA, inheritance path shown by the red arrows above, is passed from mothers to both sexes of their children, but only female children pass it on. Therefore, both men and women inherit their mitochondrial DNA from their mother’s direct matrilineal line. Men and women can both test for mitochondrial DNA, which reflects their mother’s, mother’s, mother’s mitochondrial DNA, on up the matrilineal line indefinitely.
In genealogical terms, mitochondrial DNA is perceived to be more difficult to use, so fewer people test, but I view mitochondrial DNA as exactly the opposite. Mitochondrial DNA represents an opportunity that cannot be afforded by other type of testing and isn’t any more difficult to use than autosomal.
The surname changes in each generation, but the DNA provides us with a rock-solid path to those common matrilineal ancestors, if people would simply test and upload their trees. Mitochondrial DNA has the potential to, and does OVERCOME the challenges surname changes in a way that no other tool can. The answers are written in our mitochondrial DNA along with the mitochondrial DNA of all of the people who descend from that female ancestor through all women to the current generation, which can be male or female.
When discouraging people from mitochondrial testing by telling them not to bother because it’s hard to use, the genealogical community actually perpetuates the problem. Here’s a wonderful series about how to understand and utilize mitochondrial DNA.
If EVERYONE would test their mitochondrial DNA, we would be breaking through brick walls at lightning speed. Mitochondrial DNA isn’t difficult because it’s harder to use, it’s difficult because not enough people have tested.
The surnames in autosomal lines are different from that of the tester too, yet genetic genealogists don’t hesitate for one second to take an autosomal test where they will need to build out trees to attempt to determine which line their matches connect through. The great news about mitochondrial DNA is that you already know which line the connection is through – your matrilineal line.
Mitochondrial DNA and Y DNA provide laser-sighted focus on the history of one specific line, reaching deeply back in time with no admixture from the other parent. Autosomal DNA is broad, but not deep, because it is divided in half in each generation as it’s passed from parent to child.
I wrote a series of articles, here, about mitochondrial DNA with step-by-step instructions about how to use mitochondrial DNA successfully.
Autosomal DNA, the third kind of DNA testing is the Family Finder test at Family Tree DNA, or the MyHeritageDNA, AncestryDNA and 23andMe tests which provide matches to people from all of our genealogical lines. In the graphic above, I’ve represented autosomal DNA by the broken green arrow, indicating that autosomal provides matches and links to some of the people who descend from common ancestors, but not all.
In each generation, autosomal DNA is divided in half, meaning that each person receives half of their mother’s and half of their father’s autosomal DNA. We match all of our first and second cousins, but only about 90% of our third cousins who descend from our common great-great-grandparents, in the green arrow generation. As we move further back in time generationally, we match fewer and fewer of the people who descend from common ancestors.
Therefore, I classify autosomal DNA as broad, meaning we match descendants from more than one line, but not deep, because it only positively reaches back 3 generations, often reaches back about 5 or 6 generations, but generally not more than 9 or 10 generations. The ONLY way to see back further in time than autosomal matching is Y and mitochondrial DNA.
Y and mitochondrial DNA is deep, meaning we each match only one line for each type of DNA, reaching very far back in time, but not broad. Therefore, in order to “collect” the Y and mitochondrial DNA of each of our ancestors, we need to find the appropriate cousins to test to provide us with that information.
What Can Y and Mitochondrial DNA Do for Us?
Fortunately, Y and mitochondrial DNA have the ability to help us with close relatives and matches as well as more distant history.
Y and mitochondrial DNA both have the ability to:
- Assist us genealogically to connect in recent generations utilizing surnames, trees and geography
- Facilitate identification of each lineage further back in time by utilizing full haplogroups
- Provide us with the ability to view the geographic locations of the earliest known ancestors of our matches on our Matches Map at Family Tree DNA which can provide clues as to the identity of common ancestors
- Document our ancestors’ migrations and locations where haplogroup clusters are found throughout the world
- Identify the “ethnicity” of those ancestors by haplogroups that occur only in specific portions of the world (like the Americas), or unique populations (such as Jewish people.)
- Granular personal haplogroup allowing testers to use the public Y tree and public mitochondrial tree.
Trees of Mankind and Womankind
Aside from our own personal genealogy, Y and mitochondrial DNA testing, tracking those mutations back in time, has scribed the history of the migration of mankind – and womankind – which means it tells us where our ancestors came from, and went.

On the map at left, the basic Y DNA haplogroups are shown expanding out of Africa, into the Middle East and then into Europe, Asia, the Pacific Islands and Americas. Think of haplogroups as large genetic clans, defined by mutations that group us together, and separate us apart as well.
Mitochondrial DNA haplogroups followed the same paths of course. Different locations in the world have specific haplogroups further broken down into sub-haplogroups associated with general geographic locations.
For example, Native American aboriginal people in North, Central and South America are defined by subsets of Y DNA haplogroups C and Q, and mitochondrial haplogroups A, B, C, D and X.
It’s testing by many people, citizen scientists and genealogists, people just like you, that have allowed scientists to define these haplogroups and their migration paths across the world. We still continue to discover, define and refine that pathway today. We’ve only seen the tip of the iceberg. Our knowledge is ever-changing and expanding. There’s so much more to learn. That’s why we’ve launched The Million Mito Project.
Connecting the Dots Using Mutations
How do scientists, and genealogists, connect those dots from today’s testers to their ancestors?
Mutations, called SNPs, single nucleotide polymorphisms, occur at a specific point in time and the resulting variant (mutated value) is then passed to all of the descendants of the person in whose DNA they occurred.
These haplogroup-defining mutations accumulate over time to form twigs, then branches, then the haplotree backbone as we move further back in time. On the slide below:
- Branches – green and magenta dots
- Twigs – white, yellow and grey dots
- Leaves – teal, violet (line 10) and black dots
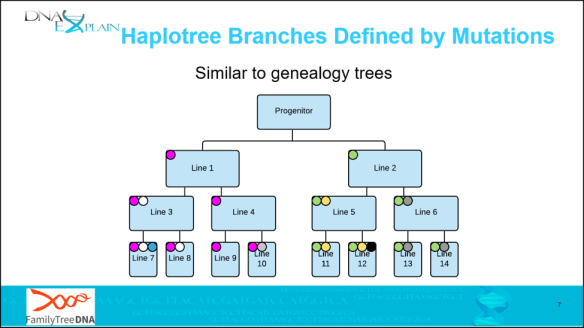
In this example, the progenitor is shown as a male, but the concept is the same for any mutation.
- The progenitor has two sons, “Line 1” and Line 2.”
- A magenta mutation occurred between the father and son in “Line 1”, and you can see that it’s passed to every descendant of “Line 1.”
- A green mutation occurred between the father and son in “Line 2”, which is also passed to all of Line 2’s descendants.
- “Line 3” had a white mutation, which was passed to his descendants.
- “Line 6” had a grey mutation which was passed to his descendants, and so forth.
In real life, mutations generally don’t accrue this rapidly, but the compressed time in this illustration makes the generational inheritance of mutations easy to see.
In both Y and mitochondrial DNA, SNPs are what form branches of the tree. In our case, the progenitor would be the trunk, Line 1 and Line 2 would be major branches, and with each succeeding SNP generation, smaller branches and twigs being created. Multiply this mutation process over hundreds and thousands of years to construct the Y tree of mankind and the mitochondrial tree of womankind.
The Explosive Expansion of the Y DNA Tree
In the past decade, great strides have been made in fleshing out the Y DNA tree.

- In 2010, only 441 branches, known as haplogroups, existed on the Y DNA tree.
- In July 2012, when the Genographic version 2 was released, the Y tree had 850 branches. The Geno 2.0 custom chip tested more than 12,000 locations on the Y chromosome with the hope of dramatically expanding the Y DNA haplotree.
- By November 2013, Family Tree DNA and the Genographic Project had jointly expanded the tree from about 850 SNPS, or haplogroups, to over 6200, of which 1200 were “terminal SNPs,” which were end-of-line SNPs in 2013. Those were the “leaves” terminating branches at that time, but most of those branches now have leaves on their twigs beneath them today.
- As of February 2020, there are more than 26,600 individual tree branches identified and named on the FamilyTreeDNA public Y DNA tree, available here, with another half million Y mutations identified, waiting for additional testers to confirm their SNP status by matching. At that time, the mutation can be given a SNP name and placed in their proper location on the Y tree, sometimes as a new leaf or occasionally, splitting an existing branch.
The key to this success has been thousands of men purchasing the Big Y test at Family Tree DNA with the hope of learning more about their paternal genealogy; first the Big Y, then the Big Y-500 and then in 2019 upgrading to the Big Y-700 with significantly increased capabilities.
Every tester can see their place on the Y block tree on their personal page, along with their matches.
Everyone, whether they have taken the Y DNA test or not can view any haplogroup’s location on the Family Tree DNA public Y DNA tree, here.
Mitochondrial DNA Tree
However, mitochondrial DNA has been neglected. The goal of the Million Mito Project is to change that.

In March 2017, FamilyTreeDNA updated to mitochondrial DNA Build 17 of the mitochondrial tree which included 5437 haplogroups extracted from just under 25,000 sequences. Family Tree DNA created an easily accessible public tree, here, complete with geographic locations for testers assigned to each haplogroup.
To date, the various mitochondrial builds have been created using GenBank submissions, but the majority of testers don’t upload their results to GenBank.
The Genographic Project and FamilyTreeDNA databases together hold more than half a million full sequence mitochondrial tests, far more than the 25,000 utilized for Build 17.
What is a Full Mitochondrial DNA Sequence?
Mitochondrial DNA is comprised of 16,569 locations in total. Initial DNA testing was expensive, so the mitochondria was divided into three regions for testing, analogous to a clock face:

- HVR1 (hypervariable region 1) – think of this area as 5 till the hour until the hour (5 minutes)
- HVR2 (hypervariable region 2) – think of this as the hour until 5 after the hour (5 minutes)
- Coding region (CR) – think of this as the space between 5 after the hour to 5 till the hour (50 minutes)
All three regions together, meaning the entire clock face, is known as the full mitochondrial sequence, or FMS.
In order to obtain a complete haplogroup designation, one must test the entire mitochondrial sequence, all 16,569 locations. The full sequence is also necessary for maximum genealogical usefulness.
Tests like 23andMe and LivingDNA provide testers with a base haplogroup as part of an autosomal test by testing a subset of approximately 1200 mitochondrial locations known to define the upper branches of the mitochondrial haplotree. I wrote about the differences, using examples, here.
There is nothing wrong with those tests, as far as they go, but they aren’t useful except as an exclusion for genealogy. In other words, if you are estimated to be haplogroup J1c, you clearly aren’t related on that line to someone that’s H1a. You may or may not be related in hundreds or thousands of years to someone else who is estimated as haplogroup J1c. You need both a full haplogroup designation, which in my case is J1c2f, and matching to make that determination. You can only receive those features by testing your full mitochondrial sequence at FamilyTreeDNA.
For mitochondrial DNA to be relevant for genealogy, or science, every location must be tested and matched to other testers. To do otherwise is analogous to having only a few of the words in your ancestor’s will and attempting to draw conclusions from only a small portion of the available data.
Full sequence mitochondrial DNA tests benefit genealogy and science too. Better yet, mitochondrial DNA gives us something to work with when we’ve exhausted all records and we have nothing else available.

Estimates today are that at least 30 million people have taken autosomal tests for genealogy, yet less than a million have taken a mitochondrial DNA test.
When autosomal testing was new, close matches were seldom found, but as the number of testers increased, it became common to find close matches of family members you didn’t realize were testing – or close relatives you don’t know. In other words, the usefulness of these tests is in direct proportion to the number of people who test.
Approximately 2% of autosomal testers have taken full mitochondrial sequence tests. Imagine how many brick walls would come crashing down if all testers, male and female, tested their mitochondrial DNA AND provided trees.
We have many examples of success stories today, even with limited testing. People discover that their ancestors were Native American, or not, Jewish, or not, African, or not. They discover their ancestor’s siblings along with breadcrumbs connecting records and people in two places as descendants of the same family.
You can read a few success stories here, here, here, here, here, here, and here – you get the idea, right?
Sometimes mitochondrial DNA is all we have when a woman’s surname is missing. But guess what – before you can be successful – you have to test. It pains me greatly to hear well-intentioned but misinformed people discouraging potential test takers.
Please add your own mitochondrial DNA success story to the comments at the end of this article so genealogists can see for themselves the power of mitochondrial DNA.
Benefits
The Million Mito Project will benefit testers in the following ways:
- Additional testers and matches
- Adding to and refining the haplotree which results in providing more useful genealogical information between matches
- Identification of and naming many new haplotree branches
- Aging and dating of the branches of the mitochondrial haplotree, enabling groups of testers to estimate when their most recent common ancestor lived
Comparing my own results to those of my closest matches, and those of individuals within the projects I administer that have authorized me to view their full sequence results, I can see that many groups of people exist that share common mutations and likely qualify to become a sub-haplogroup.
As with the Y-DNA tree, FamilyTreeDNA and the Genographic Project are in the best position, collaboratively, to combine forces to rewrite the tree of womankind. Given that 25,000 samples resulted in 5,000 haplogroups previously, I can only imagine the impact of one million testers.
Will you be part of that million?
Participation
Here’s how you can participate.

- If you tested with the Genographic Project before November of 2016, you can transfer your results to FamilyTreeDNA, for free, by clicking here, then “Upload DNA Data,” then click on “Nat Geo’s Geno 2.0 DNA,” shown below.

The Genographic version 1 and 2 results are partial, not full sequence, and after transferring you will be able to upgrade to the full sequence level.
- If you tested with the Genographic Project after November 2016, your test was run in a different lab, on a different chip platform, and you cannot transfer your results to FamilyTreeDNA.
- If you opted-in to anonymous research through the Genographic Project, your results will be included. If not, you can still participate by purchasing a full sequence test at FamilyTreeDNA and you’ll receive genealogical matching as well.
- If you tested with Genographic after November 2016 and don’t know if you opted-in to research through the Genographic project, but you would like to, please contact Dr. Miguel Vilar at mvilar@ngs.org. If you tested before November 2016, simply transfer your results to FamilyTreeDNA as described above.
- If you tested directly at Family Tree DNA at the HVR1 or HVR2 level which is called the “Plus” test, you can upgrade to the full sequence level by signing on to your account and clicking on “Full” to upgrade.

- If you’ve tested at FamilyTreeDNA at the full sequence level, there’s nothing to purchase. You’re all set – and thank you!

- To verify your testing level at FamilyTreeDNA, sign on to your FamilyTreeDNA account to verify that the “Full” icon is red.
Million Mito Genographic Transfer and Participation Summary
I’ve created a grid to summarize the three Genographic test types and how each can participate in The Million Mito Project. None of the Genographic results will be available to testers or to transfer after June 30, 2020.
| Genographic Test Type | Date | Participate in Million Mito Project? | FTDNA Transfer available (before June 30, 2020) | What Transfers? | Upgrade Needed to Full Sequence? |
| Geno 1 | 2005-2015 | Yes, via transfer | Yes | HVR1 values | Yes |
| Geno 2 | July 2012-Nov 2016 | Yes, via transfer | Yes | Partial haplogroup SNPs only | Yes |
| Geno 2 Next Generation | Nov 2016-2019 (tested through Helix) | Through Genograhic, only if opted- in to Genographic Research, otherwise test at Family Tree DNA | No | Transfer not available | Order full sequence test from FTDNA to obtain matching and other benefits |
Family Tree DNA Participation Summary
| Test Type | Participate in Million Mito Project? | Upgrade Needed to Full Sequence? |
| HVR1, HVR2 | Yes, need upgrade | Yes |
| Full Mitochondrial Sequence (FMS) | Yes | No upgrade needed |
Be One in a Million
Science needs you.
Your ancestors are waiting to be found.
Will you join us in the quest to advance science while solving the mystery of your ancestors by taking or upgrading to a full sequence mitochondrial DNA test?

Become a part of history. Click here to test, upgrade or transfer your mitochondrial results, today!
_____________________________________________________________
Disclosure
I receive a small contribution when you click on some of the links to vendors in my articles. This does NOT increase the price you pay but helps me to keep the lights on and this informational blog free for everyone. Please click on the links in the articles or to the vendors below if you are purchasing products or DNA testing.
Thank you so much.
DNA Purchases and Free Transfers
- FamilyTreeDNA – Y, mitochondrial and autosomal DNA testing
- MyHeritage DNA – ancestry autosomal DNA only, not health
- MyHeritage DNA plus Health
- MyHeritage FREE DNA file upload – transfer your results from other vendors free
- AncestryDNA – autosomal DNA only
- 23andMe Ancestry – autosomal DNA only, no Health
- 23andMe Ancestry Plus Health
- LivingDNA
Genealogy Products and Services
- MyHeritage FREE Tree Builder – genealogy software for your computer
- MyHeritage Subscription with Free Trial
- Legacy Family Tree Webinars – genealogy and DNA classes, subscription based, some free
- Legacy Family Tree Software – genealogy software for your computer
- Charting Companion – Charts and Reports to use with your genealogy software or FamilySearch
Genealogy Research
- Legacy Tree Genealogists – professional genealogy research
Fun DNA Stuff
- Celebrate DNA – customized DNA themed t-shirts, bags and other items
Discover more from DNAeXplained - Genetic Genealogy
Subscribe to get the latest posts sent to your email.


Do I understand this correctly? If I have already done a full mtDNA sequence at FamilyTreeDNA, then I need do nothing to participate in the project. Is that right?
Correct.
Thanks, that’s exactly what I wanted to know!
Hi Roberta – I have a confirmed FMS at FT-DNA, I also have two sisters but they should have the same data. It’s expensive, so should they also do mtDNA testing?Would it help the science in any way?
No, only one of you needs to take that test. Sounds like you are good.
Therefore can I assume assume that this is also true for a maternal 1st cousin?
Yes.
My LivingDNA result is J1c3j, not too far from yours. Would not such results add to the project? The ancestor is my 4g grandmother, born c 1790 in (probably) Templemore, Tipperary, Ireland, (perhaps) County Mayo.
Yes, it’s your additional mutations that will determine a new haplogroup.
Living DNA is now showing my complete Haplogroup. Even though I use my mt haplogroup from FTDNA as part of my user name, I do not think.they took it from tgat.
This is what they show for me.
“Your haplogroup is U5a2c3a”
I add my mt haplogroup to my user name at GEDmatch and MyHeritageDNA also.
I am hoping it will motivate others whose family has this haplogroup and.also who have an autosomal match to.me to contact me.
I absolutely recommend doing the full sequence mtDNA test at FTDNA. You will need to research the maternal ancestors of your close matches. It works! I found my 4th great grandparents, Edward Cook and Priscilla Baldwin of Pittsylvania Co, VA through mtDNA matching. I still do not know the first name of my 3rd great grandmother who was their daughter, but I know her children, grandchildren, her husband Martin Durham of Caswell Co, NC, and through mtDNA, her parents, sister, and brother. I am from a daughter, Milla Durham of Caswell Co, NC who.married John Lay Sr of Pittsylvania Co VA and moved to Pulaski Co, KY. They are great grandparents of my mother.The mtDNA helped me find the autosomal matches and the triangulations to her father’s side of the family.
I would not know anything about being connected to Edward Cook and Priscilla Baldwin with out mtDNA testing. My match from them does not reply, so I had to do my own research of their ancestor.
I am also researching.more of this same family now who descend from Phoebe Evans and Jesse Wells, parents of Sarsh Wells, and researching Sarah Osborne who also.connects some way to Priscilla Baldwin’s family. I believe these others frm my mt matches may may lead me to parents of Priscilla Baldwin. So mtDNA keeps paying dividends. I have several matches from Sarah Wells and Sarah Osborne and their locations and autosomal matches plus my research of their families tell me they are more of Priscilla Baldwin’s relatives. Just different lines and generations. It is exciting to see these different mt matches tie together as one big family!
Janis, I’m disppointed that we aren’t mt-DNA matches, since my full haplogroup is U5b2a2b1. My matrilineal line also goes back to Pittsylvania Co., to Agnes Frances Richardson, Rebecca Jennings, and Susannah Norris. With your encouraging words, I won’t give up trying to use mt-DNA to reach back farther on this line.
Does anyone know if this is something LivingDNA are in the process of doing? My haplogroup is still simply V at LivingDNA, whereas at FTDNA it is V-C72T!
They can’t do this with an autosomal test, so no.
Success story: my mother did both the autosomal and full mtDNA tests at FTDNA before she passed in 2015. She has only 2 exact mtDNA matches (gd = 0). The interesting thing about her matrilineal line is that it parallels one of her Mayflower lines almost perfectly up to and including our ancestress Sarah Bartlett, b ca 1678 in Plymouth Colony. Further back it deviates off into a line that goes into Nottinghamshire, England. I decided to find the place where our line intersected with those maternal lines of the exact matches. Sure enough, the other ladies’ maternal lines crossed ours at Hannah Pope Bartlett, mother of the above Sarah Bartlett.
I love it when the documents agree with the DNA!!
JoAnn – please email me at jim4bartlettsatverizondotnet. I set up and Admin an FTDNA Project for the Mayflower BARTLETTs. Thanks, Jim
There is a big hole in this project – those of us who tested with Living DNA cannot upload our haplogroup to FTDNA.
It appears that FTDNA puts profits from test-takers above the success of the project which is very sad.
Living DNA does not do either a full sequence or even HVR1 or HVR2 test. They do target testing for base haplogroups, which is not the same thing. We need all the locations, not just a few for this project. New haplogroups cannot be discovered using target testing and matching cannot occur. That decision is not about money, but about good science and consistency in the database. There a link about this in the article.
Will FTDNA allow a FMS upload from other testing companies? There is increasingly more people who tested their FMS and it would be a pity not to leverage this data.
I approached FTDNA in the past and they refused to include my external FMS sequences in the database for a small fee.
Not that I know of. Where else did you take the full sequence test? One if the issues is quality and reading the same way.
There is just 16500 letters.
I have mtDNA data from YSeq and Dante, if you want specifics…
16569, plus insertions and deletions, of course. The only data that can be used in the project is data directly from labs. That means at FamilyTreeDNA, or through the Genographic Project, or from academic papers. In other words, we can’t use files submitted directly by individuals because they may have been tampered with.
Thanks for pushing forward with this new project. I will be excited to see where it leads.
My mother’s FMS is the rare (i.e. under represented) Hg X (X2d1a). My basic understanding is that this mt Hg is often associated with North American Native populations, but my mother is of northern Italian origin (Lucca). She has only one FMS match at a GD of 2, also of Italian origin but from Sicily which we have not yet been able to connect.
At the HVR-1 level, my mom has 321 matches, but only 11 of those matches have tested FMS. Of those 11 FMS, only 3 match at X2d1a (after excluding my own test and my sister’s test) one of which is my mother’s GD2 match, so I assume that the remaining 2 are further than GD2, suggesting that there is room for more subclades beneath X2d1a.
If I had 2 wishes granted to me by the DNA Genie, they would be:
1) that testers would list their furthest known Matrilineal ancestor instead leaving that space blank or (gasp!) entering a male’s name
2) that Family Tree would offer a targeted Sale to upgrade existing HVR-1 & HVR-2 kits up to FMS.
Thanks again Roberta for all you do for the Genetic Genealogy community.
I checked the mitochondrial tree for X2d1a and I show no Native and 4 in Europe. Part of X is definately Native, but it does not appear that X2d1a is. Can you refer me to where you found information that it is Native? Thanks so much.
Lack-of-success story. I did the full sequence in 2015 because my mom was a foundling. Autosomal DNA solved the mystery of who her parents were (plus lots of records, research, tears, and great mentorship at DNAAdoption.org). What has MtDNA revealed? Nothing. We now know her matrilineal line leads to Margaret Smith of New Jersey, who married in Camden in 1880. Who were her parents, other than two people from Ireland? I have theories. Full matches at FtDNA lead to a very wonderful match with a Smith–but she has never responded to contact. The other full matches are all over the map.
The great news is that DNA continues to work for you as new people test. HVe you actively worked each match? That’s how I broke through a brick wall.
Actively for the last 4 years! Nada.
I’m very excited about this project, and I love the “inseparable” logo. How should I cite it?
For those who have tested at LivingDNA, I still don’t have a complete list of the sites they sample for haplogroup assignments. The raw data download contains only the locations where you differ from the CRS. Thus you may not know whether you were actually negative for a certain haplogroup marker or whether it wasn’t tested at all.
It’s here at Pixabay: https://pixabay.com/vectors/male-female-symbols-intertwined-311240/
I really liked it too.
Pingback: Pandemic Journal: Sanity & Strategies to Keep It | DNAeXplained – Genetic Genealogy
Well done with launching this project. I have taken your advice and tested some of my relatives to get the mitochondrial DNA of some of my other ancestors too.
On my own kit I am H16b and have 98 matches yet none with a genetic distance of 0. Am I right to say that if lots more people test that I could be allocated as part of a newer branch further downstream (and thus have closer matches)?
Possibly. Do you know if you have a heteroplasmy?
No, I don’t have one.
You may be pleased to hear we have been explaining Mitochrondial DNA more recently (I was also planning to do a class on it until Covid 19 came along!). We have 10 people awaiting full sequence results in our North of Ireland project group at the moment so it is good to see the interest growing here.
Wonderful.
Roberta, I have a heteroplasmy. FMS results H1, almost 800 matches with GD1, GD2, GD3.
I had my maternal cousin test FMS. She, too, is H1, NO heteroplasmy. just as many matches with 3 exact, GD0.
I expected her to be GD1 with me, but to my chagrin, she is GD2 !
FTDNA’s explanation is that heteroplasmy counts for 2 differences. No further explanation.
So, I’m still wondering where that leaves all of my other matches. Example, shouldn’t GD1s be GD3?
Have you an explanation? Thanks for any understanding you can add.
I would need to look at your results.
Would your Quick Consult fee cover my question? If a heteroplasmy counts for 2 differences, how can I have any GD1 matches, which I do — dozens?
They might have the same heteroplasmy. I can’t see their results so a consult won’t help you.
I did my MtdDNA at Family Tree DNA but I need to upgrade to Full Sequence. I just spent a half hour at their site and I cannot figure out how to do that. Is it not possible? Do I have to do another swab?
On your mtdna part of your personal page, look to the right. You’ll see Plus and Full. Click on full to upgrade.
Pingback: Friday's Family History Finds | Empty Branches on the Family Tree
You’ve inspired me, Roberta. I’ve been wanting to do it and you just gave me another good reason. Used your link 🙂 and placed my order today.
Thank you!
UPDATE – Logsdon – Carman – LaRue – Carmen – Duwys Matrilineal Line
Your call for MTDNA testing inspired me to start contacting women who could Share MTDNA group H1u1 with me. I only contacted a few a first to see if I could get at least one other person sharing this line to test.
One challenge for creating a personal MTDNA project is naming it. Our paternal line Y-DNA projects are so simply named by the Surname and maybe a date and location, to gather testers. I actually heard on a genealogy Podcast ( I won’t name) the guest speaker advising against getting a MTDNA test as the results were so hard to use, women’s names keep changing !!!! I can imagine his family tree, he only researches male lines. We women genealogists are not so faint of heart – right !!!
The first DNA test I did was a MTDNA test. I come from a long line of awesome women, pioneers, tough, resilient and builders of this country. In ordering a MTDNA test, I picked just the right one. Reading and learning about MTDNA is fascinating and more so when I have my matrilineal line to study.
The result of my emails to potential testers ????
A BIG DRUMROLL – I now have one full MT-DNA test match at FTDNA. This tester and I are 5th cousins once removed and share common ancestors William Logsdon (1773) – Mary Elizabeth Carman (1774).
We have one exception in our match, I have an extra mutation in the Coding Region. I would give the number, but I don’t quite know what I should be careful about sharing. Apparently. somewhere in my line through 5 generations the MTDNA picked up a new mutation. My match did not lose a mutation as her record shows no deletions.
Soon I will reach out to other potential testers. I am lucky as my match is also eager to pursue our project. We only have so many potential testers to ask, so we are trying to figure out the most convincing way to reach out to them. I think the best would be to put together a web page about our matrilineal ancestors with reasons it is so important to test and test now. For one, we could easily lose all potential testers. Our numbers are not large.
BTW, my one match and I also share autosomal and X DNA which I think implies that our matching segment is from Mary Elizabeth Logsdon and not William.
At any rate, thanks so much for providing so much MTDNA information on your blog. I would be lost without it. I particularly liked the article where you talk about MTDNA combined with X-DNA, maybe you could add a link to that article on your 5 lessons page. Also, almost without saying, all your articles and examples of genealogical research are invaluable (an understatement, but what else to say).
Regards,
Linda Price
Regarding sharing information about your “extra” mutation, there are a few mutations with medical implications. You can review a list of such mutations at https://mitomap.org/MITOMAP. The list does include lots of mutations that are single case observations, and the presence of the mutation could be just a coincidence.
The word “extra” may be misleading. It is a mutation that is not used to define a haplogroup, so it is confined to a smaller lineage. It is not an insertion. It is a SNP where one base (A or C or G or T) is replaced by another one. Your match at FTDNA would retain the ancestral value and would not show a deletion in her record.
I write custom reports for people who want to delve into the details.
https://isogg.org/wiki/Custom_mitochondrial_DNA_reports
Linda I too am H1u1. I live in Melbourne Australia. my earliest known female ancestor is c1830 in Co Tyrone or Donegal, Ireland. I have a public tree on Ancestry “Chrissy2712”.
Hello ~ I have a recommendation for FTDNA and for you. Perhaps you might be able to make this recommendation if you should find it a good idea. First, a little background to get to my point.
A few years back I transferred mine and my grandmother’s Autosomal DNA over to FTDNA. My grandmother is 91 years old and having her participation has been a gold mine. She has way more matches than myself and of course, her dna goes back a much further reaching into family branches lost to me. Her family is primarily of colonial ancestors, so it is very helpful to have her dna to prove and disprove family lines.
I did my dna on 23&Me, and what I found most interesting there was my mtdna haplogroup (I4a). It’s a bit more rare than average and after time I thought I need to do the full sequence as it might open more doors in my research for my maternal line. The furthest I can go back is my 6th great grandmother born in 1797 (Dorcas Purnell). The amount of research I’ve done on Dorcas’s father has been deeply extensive and I’ve discovered things no one else had on him, but I can not find the name of my 7th maternal great grandmother. There is erroneous info as to who she was, but no one seems to pay mind to the fact that it’s not correct. Based on clues and names given to Dorcas’s siblings (a marriage of one sister to a Bounds and another sister named Ann Dykes Purnell) I had theorized a link to this Bounds family. Thrulines on AncestryDNA for my grandmother is proving a solid link to the theory I had on a John Bounds and his wife Mary being parents of my unknown 7th great grandmother. John Bounds mother was Ann Dykes which we find in the Purnell family. Interesting enough, I have three people on FTDNA Mtdna full genetic match (distance of 2 and 3) that show THIS Ann Dykes who was married to a James Bound as their furthest maternal ancestor. I’d say I’m on the right track, but it can make things even more confusing. Furthermore, I show that two other matches on my MTDNA FTDNA, with genetic distance of 2 and 3, are people that on my grandmother’s family finder, but not mine. Again, it raises more questions than answers, but hey in theory it’s all good.
My recommendation for FTDNA is for them to somehow allow you to link your MTDNA results with any other accounts you might manage that have to do with paternal or maternal results. If someones mother/father or grandmother/grandfather decides to do their dna then would be extremely helpful to add the appropriate family finder results to one’s Mtdna or Y results for advanced matches reports.
If I could link my grandmother’s family finder results to my Mtdna full sequence results for reporting only, I would see some alignment with HVR1/HVR2/FMS and Family Finder with names. Her dna allows a deeper reach of matches and this would be incredibly helpful.
Currently, I have to try to take each name and place it into her family finder. It is a lot of back and forth, not to mention getting logged out of one account and having to relog into the other account.
I am very fortunate to have a lot of matches and a lot of EXACT matches at 0. Many are in Ireland or can trace back to Ireland with recent great grandmothers. I would love to be able to finally break through my wall because I want to find my immigrant maternal great grandmother someday. I’m guessing she is probably Irish, but I don’t want to assume.
Thank you for your time and well, I can’t tell you how much I have appreciated your blog all these years. I have a natural knack at researching and I find it amusing that I always find out later that I am doing what you are already suggesting. I feel like Columbo in genetic genealogy. All my best! Be well.
That’s a good idea. In the mean time, Genetic Affairs has the rule-based clustering that allows you to combine kits and includes both Y, mtDNA and autosomal in the analysis.
Pingback: Bryan Sykes Finally Meets Eve’s 7 Daughters | DNAeXplained – Genetic Genealogy
Could you clarify, please if we know for certain whether the
”f” mutation in J1c2f actually occurred prior to, say, the ”g” mutation in J1c2g?
Does J1c2g also have the ”f” mutation or do the similarities between J1c2f
and J1c2g actually end at J1c2? Thank you. Best wishes for 2021.
The order of g and f are simply the naming order based in then they were discovered. Their common ancestor ends at J1c2.
Having been earmarked for a potential new subclade ever since the haplotree was last updated it’s exciting to hear that work is underway for a new update.
My half 4th cousin who shares my matrilineal 3GGM agreed to test for me and her sample is en route to FTDNA.
The results are likely to be too late for the first paper from this project but I am excited about the outcome.
I can’t talk about the first paper yet, but it does not adjust the entire tree. It’s an important discovery.
I’m confused, perhaps you could help.
My mtDNA is showing up as Jlc3b. My brother is Jlc3, my daughters are Jlc3, my granddaughters are Jlc3’s. Why am I Jlc3b? Also, would Jlc3 matches be different than Jlc3b.
Where did you test? If you didn’t test at FTDNA, you’re only receiving partial haplogroups. It looks like maybe you did and the rest didn’t?
When can we expect updates based on this project to current haplogroups at Family Tree DNA? YFull defined a new haplogroup for my father and his match, but I guess it is not an official haplogroup.
We are nearing completion of the first paper.
I am working on a collaborative wiki tracing the Y-DNA and mtDNA of Mayflower passengers, their spouses, and their sons in-laws. This project connect DNA to documented trees and hopes to expand the knowledge base. The link to this wiki is: https://mayflowerDNA.org
I’m all in on this!!!
I’m U5b1b1-T16192C! at FTDNA. At YFull this is U5b1b1-b*. I’m the only one with mutation G228C. Will that mutation eventually lead to a new subclade? And is the YFull hg the updated subclade naming? Thank you.
There is no updated haplogroup naming yet. And new haplogroups can’t be named with just 1 person.
Dear Roberta,
I found the contact to you via FTDNA. I find the Millon Mito project very exciting, so I would also like to contribute to it. So far I have done two tests and got the following results:
23an ME => H1q
Nebula WSG => H1q3
I have already transferred the 23andME data to FTDNA.
How can I support your project?
Via classical genealogy I could prove my matarnal line closed until about 1650. What additional value can I get from the mtDNA?
best regards
Wilfried
You’ll need to order the mtFull test which tests more extensively, plus you receive your mtDNA matches. Thank you.
Does that mean the terminal haplogroup from the Nebula test is wrong? I thought so far that is not possible at all. What is the real reason why FTDNA does not want to start with these results. It would be a pity to do without them or?
The reason I don’t recommend whole genome testing is because you don’t have the matching capabilities provided by the genealogy vendors. You paid for a test with a vendor with no matching database. You got what you paid for.
Your Nebula WGS would almost certainly be correct and complete. It might even be more sensitive to “heteroplasmy” (a mixture of mtDNA molecules with and without a certain mutation), but I don’t have any concrete examples. I thought Nebula had some sort of agreement with FTDNA, but I’m not certain what the current status is.
Regarding the mtDNA Haplogroup 23andMe assigns to folks: Autosomal tests such as 23andMe (as well as AncestryDNA and even FTDNA’s Family Finder test) use a special chip to detect a select number of SNPs. The process of creating these SNPs is not 100% perfect so on occasion, the Haplogroup assigned to an individual may not be correct. This affects their Y-DNA assignments much more than mtDNA assignments (simply due to the size difference between the Y Chromosome and the mtDNA), but it is possible for errors to occur in mtDNA assignment as well.