Update: The WTY has been superceded by the Big Y test, but I’m leaving this article for historical continuity.
What the heck is WTY….and why do I care?
One of the reasons I started a blog is to continue what I do for my clients when I write their DNA reports. I make DNA understandable and fun for the normal air-breathing genealogist.
The past few days has been a whirlwind of information and announcements, some which tend to leave folks who don’t have a lot of experience in the dust.
For that, I do apologize. However, I’d like to tackle a much easier topic now, and that’s the WTY test. What is it and why is it so important?
WTY is short for Walk the Y, as in walk down the Y chromosome.
The tests we all order and love, at Family Tree DNA, that would be the 12, 25, 37, 67 and 111 marker tests, tell us about genealogy – who we are related to in the past several hundred years.
Deeper ancestry, anthropological in nature, a line I draw about the time when surnames were being adopted, is different and little information of that nature is exposed by the STR (short tandem repeat) genealogy markers.
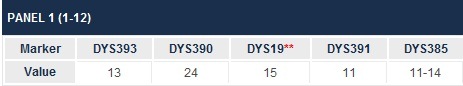
By the way, short tandem repeat means those locations in our DNA that are prone to develop repeated sequences. Think of them as genetic stutters. They are important to us as genealogists, because on the Y chromosome, we count the number of those stutters and that is the marker value reported.
For example, below, we see that for marker 393, we have a value of 13. That means there were 13 repeats of the same sequence. Obviously, combining all of these sequences, or marker values, together creates our own genealogical genetic profile or fingerprint. This, of course, is what we use to compare to others to see whom we match.
However, deep ancestry, identified by our haplogroup, is determined by a different kind of mutation, called a SNP, a single nucleotide polymorphism.
These are mutations that happen in only one location, and they are considered to be once in the lifetime of man mutations. In actuality, these mutations sometimes happen independently in different haplogroups, but the cumulative sequence of SNP mutations defines our haplogroup.
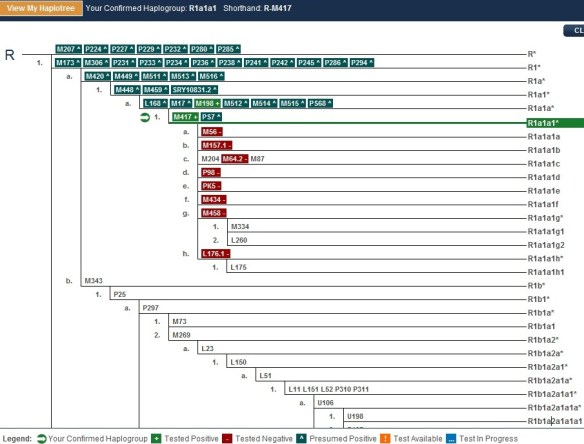
You can see, for example, below, a haplotree from a Family Tree DNA client’s results page.
This person tested positive for the light green SNP, M417. The plus means that they have this specific mutation. In his case, this is his terminal SNP, meaning the one furthest down the tree that defines his haplogroup, as we know it today. That would be R1a1a1.
The SNPs shown in red, below M417 are ones that he has also been tested for, but does not have, so he knows he is not a member of those haplogroups. These are shown with a minus sign, such as M56-.
Now for the problem that WTY has been helping to solve.
If your STR markers take you back about 500 years, in round numbers, and your haplogroup tells you where your ancestors were between 3000 and 4500 years ago, in this case, where were they in-between? What were they doing? Where did they live and how did they get from where they were 4500 years ago to where you find them 300 or 400 years ago, if you’re a lucky genealogist and can go back that far?
There is a significant gap in the timeframe between STR genealogy markers and haplogroup SNP markers. Finding additional SNPs will eventually close the gap between STR genealogy markers and haplogroups. We will have a complete timeline of our ancestors. In some cases, we’re even finding family-specific SNPs, known as “personal SNPs.” How cool is that? A new haplogroup is born in your family!
Did you notice on the tree above that some of the SNP markers begin with L? Every SNP discovered is prefaced with a letter that tells people which lab or university discovered the SNP. The L SNPs have all been discovered at Family Tree DNA’s Genomics Lab in Houston, Texas, run by Thomas Krahn. They are the product of the WTY discovery process.
When there is reason to believe that a SNP might be lurking undiscovered in the DNA of a person or a group, then the WTY becomes an option. Generally, the clue would be STR markers that are significantly different than any previously seen, or part of a small and quite unusual cluster.
Today, we test all of the known downstream SNPS, the ones in red above, and then if none are found, we would apply to Family Tree DNA to do a WTY test. This test is quite labor intensive. In essence, they manually look at between 450,000 and 500,000 positions to see if they spy any new mutations.
If they do, they begin the SNP naming process and the process of getting the SNP officially added onto the tree. You can see the most current haplotree (Y SNP tree) at the ISOGG site. Because of the long naming and authentication process, sometimes trees at different locations aren’t quite in sync. The ISOGG tree, maintained by volunteer genetic genealogists, has become what most people look to and use as the gold standard today.
In any case, this process is how new SNPs are discovered. The Geno 2.0 project includes 12,000 SNPs for the Y chromosome, an exponential growth from the current 862, or so. At least some of these SNPs were discovered at Family Tree DNA, as a result of savvy project administrators and others who are familiar enough with DNA results to suspect that a new SNP might exist, and who advocated with the tester and Family Tree DNA for WTY testing.
Hopefully, you now understand better about the WTY and why WTY tests are so critically important.
How might you know if you or a family member is a good candidate?
If you have tested to 67 or more markers and have no matches, you may be a candidate. You would need to do a deep clade test, which tests all relevant downstream SNPS at this point. In the past this has been the Deep Clade test, but today it would be the Geno 2.0 test. If you think you might be a candidate, you’ll want to work with your haplogroup administrator to see if there are any experimental SNPS to test for after the deep clade/Geno 2.0 is completed.
The WTY is the perfect example of collaborative citizen science. Participants fund part of the testing, haplogroup administrators identify good candidates, Family Tree DNA underwrites part of the testing fee and of course performs the test, and everyone benefits. Before you know it, you’ve got 12,000 new SNPs combined with new technology that promises to do more than we’ve ever dared dream before!!!
______________________________________________________________
Disclosure
I receive a small contribution when you click on some of the links to vendors in my articles. This does NOT increase the price you pay but helps me to keep the lights on and this informational blog free for everyone. Please click on the links in the articles or to the vendors below if you are purchasing products or DNA testing.
Thank you so much.
DNA Purchases and Free Transfers
- Family Tree DNA
- MyHeritage DNA only
- MyHeritage DNA plus Health
- MyHeritage FREE DNA file upload
- AncestryDNA
- 23andMe Ancestry
- 23andMe Ancestry Plus Health
- LivingDNA
Genealogy Services
Genealogy Research
- Legacy Tree Genealogists for genealogy research


Hello,
I know little-to-nothing about genetics, so please forgive my obvious ignorance. I do not understand why a 67-marker study would be a better qualifier for the WTY process than, for example, a 37-marker study with no matches. Am I wrong to conclude that there is “0” chance for matches at 67 markers if one has “0” matches at 37 markers? Since the widest “field-of-view”, to borrow an optical allusion, is inherent in the lower marker studies, and, thus, more likely to produce matches than the higher-count marker studies, what new information would the higher-count marker study produce that is relevant to matches?
Jacob
FTDNA J2, 37-marker study with no matches higher than 12 markers
Because of the way mutations sometimes fall, some poeple do actually have matches at 67 or 111 markers when they don’t at lower levels. For example, if you have more than 1 mutation in the first 12 markers, you won’t show as a match there, but with 2 mutations you will show as a match at 25 markers. It’s all in how the mutations fall in the panels.
Roberta,
Thank you for your prompt reply. Would one be able to discern from looking at the 37-marker study whether, or not, it would be advisable to order the higher studies? If so, who would be available to perform this evaluation?
Jacob
Generally, the administrator of your haplogroup project would be who you would work with. Have you joined the haplogroup projects that are appropriate for your results?
Thanks Roberta! I always enjoy reading your posts- no matter how many articles I read elsewhere trying to learn more, yours are always helpful in understanding further. You break it down for the newbies quite nicely. : )
My father’s group is going through WTY testing right now. FTDNA has found something “different” about his group, but they haven’t let us know fully what’s going on yet…….I think they found some new SNPs, but there was a communication breakdown of some sort- we had a change in contact at FTDNA for our group and things went kind of awry……
It’s exciting to be a part of scientific discovery isn’t it!
Hi Roberta,
Do you think you can “fact check” with FTDNA this statement you made:
“You would need to do a deep clade test, which tests all relevant downstream SNPS at this point.”
The “R” FTDNA Y-Haplotree is not as current as the Gold Standard and Carefully Vetted “R” ISOGG Y-Haplotree ( http://www.isogg.org/tree/ISOGG_HapgrpR.html )
A case in point is my presently Terminal Y-SNP of R1b-L371 (Wales) http://tech.groups.yahoo.com/group/RL371/
R1b-L371 was added by ISOGG in September 2011 … but I believe FTDNA still does not test for it in their $139 R Deep Clade Test. One can test for it as an individual Y-SNP at FTDNA for $29 to see if they are L371+ or L371- I am L371+ and of Paternal Welsh Origins. My Jones line descended out of the R1b-L371 Griffith line and other Welsh lines going back to the R1b-L371 Founder in Wales circa 1000 AD to 1070 AD.
So, I am pretty sure the Current FTDNA Deep Clade does not include “all” Current or “all” Pending ISOGG Y-DNA Haplogroup SNPs at this time. This price was raised this year and the R only Deep Clade Tests now costs $139 versus the new and better valued Geno 2.0 at $199.95
Also, I have checked that R1b-L371 is indeed included in the Geno 2.0 test as it was ISOGG added prior to the November 2011 Geno 2.0 cutoff. So, any Y-SNPS discovered in the 1 year period from 11/2011 to 11/2012 will not be tested for on the Geno 2.0 Illumina Chip.
Instead of taking an individual SNP test OR the Deep Clade Test OR the Geno 2.0 Test, many can look at their Y-STR values which are often very predictive of a certain Y-SNP. For example, the 100% accurate R1b-L371 Y-STR Signature is DYS448=17; DYS456=14; DYS450=10.
Failing all the above methods then the very costly approach with the FTDNA “Walk Through the Y Test (WTY)” can be considered after discussing it in detail with the appropriate FTDNA Group Admins or an ISOGG expert in this area such as David Reynolds for the “R” Y-Tree.
In addition, there are DNA analysis and interpretation methods to correlate Y-SNPs and Y-STR Signatures with various Autosomal SNP Segments or Individual Autosomal SNPs which serve as AIMs – Ancestry Informative Markers.
The POBI Research Group will be doing this in the British Isles. http://sse.royalsociety.org/2012/exhibits/genetic-maps/ http://www.forumbiodiversity.com/showthread.php?t=21518
George
Hi George,
Much of what you’ve said is absolutely accurate. This was written as a primer for folks. I did refer them to their haplogroup project administrator. They would be the ones to be closely aware of the new and pending SNPs relevant to the haplogroup.
In the past, the Deep Clade test was required. You’re right that in the future, the Geno 2.0 would replace the deep clade. That info was included a few days ago by Bennett in the update I provided. In essence the deep clade has become obsolete. The relevant point here is that before proceeding with the WTY, one wants to be sure they have availed themselves of all of the appropriate standard tests that test existing known SNPs. The WTY is not appropriate or necessary for everyone. Thank heavens for our haplogroup administrators who guide people through this process, which is different for and varies with each haplogroup.
And the POBI project promises to be very, VERY interesting.
Roberta
” In essence the deep clade has become obsolete. ”
I was about to ask if that was the case 🙂 I’ve done a couple of single SNP marker tests beyond the deep clade a while back, but neither Y (G2a3b1a4) or mtDNA (U5b3.. h?) haplogroups fit well, so going to wait for geno2.0 and see if that helps
Pingback: Family Tree DNA New YouTube Channel | DNAeXplained – Genetic Genealogy
Pingback: Family Tree DNA Conference 2012 – Native American Focus Meeting | DNAeXplained – Genetic Genealogy
Pingback: Family Tree DNA Conference 2012 – Native American Focus Meeting | Native Heritage Project
I used the national geographic test kit and found out I have the m417 marker. I am interested in finding out more. WHAT do you suggest in the way of tests to find out more.
Be sure to transfer your information from Nat Geo to Family Tree DNA. It’s free. Then join the appropriate haplogroup project, which in your case would be R1a. Then ask the admins what they would recommend based on your individuals markers and any groups clusters you fall into.
Pingback: DNAeXplain Archives – Historical or Obsolete Articles | DNAeXplained – Genetic Genealogy