There has been recent discussion and confusion about the difference between pedigree collapse and endogamy.
Let’s take a look at the similarities and differences and what it means to genealogists.
Pedigree Collapse
Pedigree collapse occurs when the same person/people appear in your tree multiple times as ancestors.
In this example, you can see that John Smith and Mary Johnson appear twice, which of course means the ancestors further back in time in those lines all appear at least twice too.
Genetically speaking, our tester, Tester Smith, could be expected to inherit more of the DNA of John Smith and Mary Johnson because they are receiving an infusion of their DNA from both sides of their tree.
Each parent provides 50% of their respective DNA to each child, but contribute different pieces of their DNA to different children.
Each grandparent normally contributes approximately 25% to each grandchild, although it may be slightly more or less. Each great-grandparent contributes about 12.5% to each great-grandchild.
However, since John Smith appears twice in Tester Smith’s tree as a great-grandparent, John Smith would be expected to contribute approximately 12.5% of his DNA times 2 to Tester Smith. This means that approximately 25% of Tester Smith’s DNA descends from John Smith. The same is true for John Smith’s wife, Mary Johnson.
Let’s look at how this affects our chromosomes and matching.
Chromosome Perspective with No Pedigree Collapse
First, let’s look at a situation where there’s no pedigree collapse. Chromosome 22 has about 72 cM of DNA that is being compared for genealogy, so let’s use that chromosome for our example, with chromosome 22 being representative of all other chromosomes (except the X chromosome.)
If each grandparent contributes one fourth of each person’s DNA, then our tester’s mother would have received approximately 25% of her DNA from her 4 grandparents, respectively, or 18 cM from each grandparent.
For purposes of these examples, I’m going to use the 25% average amount of DNA inherited for each grandparent, but you can read more specifics here and here, if you’re interested.
In this example with no pedigree collapse, you can see that our tester received 9 cM or 12.5% of each of their great-grandparents’ DNA. The great-grandparents’ DNA combined in the grandparents and then Tester’s parents such that Tester received 18 cM or 25% of their DNA from each grandparent and 9 cM or 12.5% from each great-grandparent.
Note that Tester Smith received one 9 cM piece of each color of his 8 grandparents’ colored DNA. It’s easy to visualize inheritance this way.
Chromosome Perspective with Pedigree Collapse
Our second example shows pedigree collapse with John Smith and Mary Johnson being present as great-grandparents twice.
Note that Tester Smith inherited a segment of John Smith’s red DNA from their mother and one from their father. Tester also inherited one segment of Mary Johnson’s yellow DNA from each parent.
In this situation, the red DNA segment inherited by Tester Smith’s father from John Smith and the red DNA segment inherited from Tester’s mother from John Smith could potentially be:
- The same DNA segment contributed by John Smith to both of his children, George Smith and Fred Smith, meaning those segments will match entirely.
- Partially the same DNA segments meaning that some of John Smith’s DNA that Tester Smith inherited from his parents will match each other and some won’t.
- Entirely different DNA segments meaning that although the DNA was inherited from John Smith in both cases, his children, George Smith and Fred Smith inherited different pieces of John Smith’s DNA. That DNA was passed through George Smith and Fred Smith’s children to Tester Smith. Even though both segments inherited by Tester descended from John Smith originally, they don’t match because they were different segments to begin with.
Tester Smith will inherit approximately half of the DNA from John Smith that his parents received, so their red segments of DNA could be exactly the same, partially the same, or completely different.
Since I am showing the red segments in different positions on the chromosomes, we’ll presume that the positions shown indicate chromosome location (addresses.) Since the red DNA is not in the same location on the mother’s and father’s chromosomes, the DNA from John Smith inherited by Tester from his parents are different segments.
Tester will have inherited 18 cM total from John Smith and 18 cM total from Mary Johnson (using averages). In this illustration, the red and yellow segments, respectively are two separate 9 cM segments. If by chance those two red (or yellow) segments had been inherited in adjacent locations, they would match as one 18 cM segment – even though they were really two separate segments inherited through two different parents. The phenomenon where segments from common ancestors joining each other again in descendants causes relationship predictions to be closer than the actual relationships.
Said another way, even though Tester Smith inherited 25% of John Smith’s DNA, John Smith is still a great-grandparent, albeit twice, not a grandparent even though vendors would predict someone with 25% shared DNA as a grandparent.
Of course, each generation further back in the tree means that the amount of DNA inherited from each ancestor is cut in half, so the effects of pedigree collapse become less pronounced the further back in time the collapse occurs.
Looking at our example, if John Smith and Mary Johnson were duplicated in Tester’s tree another generation further removed, Tester would inherit 6.25% times two from John Smith, or a total of 12.5% of his DNA, and the same from Mary Johnson. Another generation back in time, 6.25% total. Eventually, many of those segments will disappear entirely due to loss during recombination, so distant pedigree collapse is not necessarily discernable in this way.
To summarize, pedigree collapse occurs in a genealogical timeframe, meaning that you could at least potentially identify the ancestors who are duplicated in your tree. If you know where in your tree the duplication occurs, you can calculate the expected amount of DNA that you will inherit (assuming an exact 50% inheritance/recombination rate in each generation) from each of those ancestors.
Endogamy
Endogamy is different. Instead of one person or a pair of ancestors who are duplicated, testers will have no immediate ancestors who are the same in their tree, but they will have many historical ancestors who are identical.
Endogamy most often occurs in closed communities where out-marriage is either highly discouraged or impossible. Common examples include Jewish populations, especially in Europe with the Ashkenazi, Native Americans, Finnish people, Acadians, Amish, Mennonite and Brethren communities. Of course, there are many more.
These communities often married only within their own community for many generations. Each community member shares the DNA of many common ancestors from long ago.
In this example, the DNA from distant common ancestors is handed down to the parents from the grandparents, but the ancestral segments are shown in small pieces. I used 4.5 cM as the segment size, but endogamous samples have many small, fragmented segments below that threshold.
“Small” segments for purposes of this discussion are those below the 6 cM minimum vendor matching/viewing threshold of FamilyTreeDNA, MyHeritage and 23andMe. Ancestry’s minimum match threshold is 8 cM. The take-away here is that none of those individual 4.5 cM segments would match between testers at any of those vendors because they are below all vendor’s thresholds.
The red arrows point to small segments where the mother and father both inherited small pieces of the same identical DNA from the same distant ancestors. Our tester will receive the pink and red DNA segment from both parents, because there is nothing else in that location for them to inherit.
The green arrows show examples of identical by chance matches where the yellow and red DNA, respectively, is not handed down from one parent. Instead, the two yellow and two red segments abut and are joined to form one 9 cM segment where two individual 4.5 cM segments converge – one inherited from the mother and one from the father.
This, of course, is the definition of identical by chance (IBC) where the DNA from two parents just happens to align in such a way that the tester matches another person. However, in ADDITION to being IBC, those two smaller segments just happened to be from common earlier ancestors in an endogamous population. Because endogamous populations have a limited amount of available DNA, it’s much more common to have small segments that match in descendants – and sometimes recombine to match in larger segments too.
In this case, the DNA of unknown distant ancestors just happened to be handed down and aligned adjacent to each other.
Our tester will match to anyone else who just happens to have inherited those two small ancestral DNA segments in the same location from that same population. When the original number of ancestors is limited, so are the number of DNA segments available for inheritance, and it’s very common for random ancestral segments to align in this way. Think of each ancestor’s tiny DNA segments salting a bowl of soup. You’re going to get some in every spoonful.
If those two adjacent 4.5 yellow segments are passed down together to the next generation, they add up to 9 cM, so will be considered a match to another person who inherited those same two adjacent 4.5 cM segments from that yellow ancestor – even though that unknown yellow ancestor could have lived ages ago – long before the possibility of genealogical matches. When no new DNA is introduced into populations, the only DNA available to be passed to the next generation is the ancestral DNA that has been salting the same pot of soup for generations.
This is exactly why we see the following situations in highly endogamous populations:
- Many matches at lower cM levels due to identical by change recombination
- Many small segments in common below vendor match thresholds
- Significantly more smaller segment matches than non-endogamous individuals due to the historical ancestral DNA being passed and recombined from descendant to descendant.
A fully endogamous individual from the Ashkenazi population often has 4 or 5 times as many matches, or more, than non-endogamous individuals.
Conversely, some fully endogamous individuals from populations that have not tested many people will have very few matches, but may not be able to identify their genealogical relationship with any of their matches.
Segments Compared
In the article Concepts – Endogamy and DNA Segments, I provided several real-life examples of how endogamy affects DNA matches.
FamilyTreeDNA’s most recent matching update, among other things, has:
- Removed the segments below 6 cM from the DNA match totals
- Developed a new technique to determine and remove many identical by chance (IBC) matches
- Fully imputed all transfer kits from other vendors (yay!,) meaning that early transfers who did not previously have distant matches now do
- Recalculated everyone’s matches based on all of the above
- Developed an improved relationship prediction algorithm
- Re-predicted everyone’s relationships
While these changes benefit everyone, they provide huge benefits to people with high numbers of matches due to endogamy.
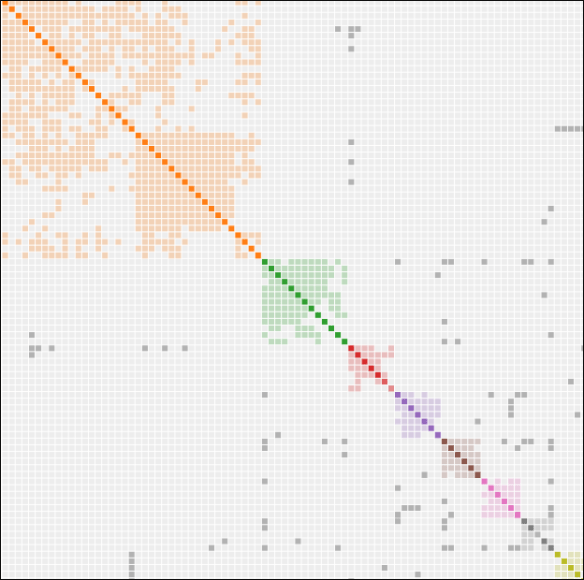
In this chart from the earlier article, you can see individuals predicted to the same relationship level, with segments as small as 1 cM showing, although matching never occurred at this level:
- Non-endogamous matches at left
- Jewish matches in the center
- Native American matches at right.
The chromosomes of the Jewish and Native people look polka dotted by comparison to the non-endogamous people. All of their matching segments are shorter then the non-endogamous group at the same predicted level, because all of the small segments were included in the relationship prediction calculations.
The removal of segments below 6 cM at FamilyTreeDNA improves accuracy and relationship predictions for everyone. A white paper will be available soon describing their new techniques.
Population Genetics
While endogamous matches are frustrating for genealogists due to both the high number of matches and the difficulty identifying common pedigree ancestors, endogamous matches are very useful in another way.
Looking at our endogamous example again, let’s say our tester is entirely Jewish, with no admixture.
Our tester has a child with a partner who is entirely Asian, with no admixture. The DNA of these two populations does not fit the same genetic pattern.
In this example, the Asian person’s DNA is chartreuse green (for simplicity.) The Jewish DNA in the child has been divided in half, losing all of the army green, bright blue, and light blue segments, along with part of the tan, grey and yellow segments. Notice that the child still has two yellow segments and two red ones.
Population geneticists look for distinct patterns among populations of people who have lived exclusively together, in close proximity, or mixed often for tell-tale genetic signals where high frequencies of certain DNA patterns, or colors here, appear. Think of an island like Australia or New Zealand where there were no new populations available.
Those telltale small DNA segments, below matching thresholds, signal membership in or a genetic affiliation with that population. Of course, not all populations are quite as distinctive as the Jewish or Aussie/NZ populations. Some populations have not been isolated as long or more admixture has taken place. Think about Europe and those fluid borders.
Still, the signal of the founding populations is present for several generations, and sometimes longer if the testers ancestors were from the same population or region of the world and those identifying segments have been preserved during genetic recombination.
This individual’s ethnicity or populations would likely be predicted at or near 50% Jewish and 50% Asian. Those populations have been separated for tens of thousands of years and are relatively easy, genetically, to tell apart.
However, ascertaining between France and Germany is another matter altogether.
Real Life Examples
In the “picture is worth a thousand words” category, let’s take a look at some visuals.
Genetic Affairs has developed autocluster technology which I’ve written about several times. In the introductory article AutoClustering by Genetic Affairs, I provided examples of “normal” non-endogamous autoclusters.
A non-endogamous individual where other people from their family lines have tested would show several separate clusters. Individuals included in the same colored cluster match each other. Those clusters represent different ancestors or ancestral couples. For the most part, the people in individual clusters don’t match other clusters, although some will as the smaller clusters tend to represent generations further back in time. The people who match two clusters are shown by grey cells.
On the other hand, people who descend from an entirely endogamous population pretty much have one large interrelated square, not neatly arranged descending colored blocks.
My mother’s great-grandfather is Acadian, a highly endogamous population. She has no known pedigree collapse outside of the Acadian population. However, the Acadian population has substantial pedigree collapse meaning that most of her matches would have substantial pedigree collapse. All Acadians share the same founding ancestors from the early 1600s.
As researchers, we are fortunate to have meticulous Catholic church and tax records maintained by the Acadians. Other genealogists aren’t nearly as fortunate and therefore can’t necessarily differentiate between endogamy and pedigree collapse or a combination of both.
Mother would have inherited about 12.5% of her DNA from Antoine (Anthony Lord) Lore.
Mom’s orange Acadian cluster at upper left is oversized, much larger than her next cluster, and you can see that many orange-cluster people are related to each other. Mom has more Acadian matches than would normally account for 12.5% of her matches at the threshold used to generate the autocluster. These proportionally oversized autoclusters are the hallmark of endogamy.
One generation further downstream, my Acadian cluster, which accounts for 6.25% of my DNA is still my largest cluster, shown below, NOT clusters from my four grandparents as might be expected.
However, my Acadian cluster isn’t nearly as large as my mother’s, illustrating just how much was lost through recombination in one generation.
My friend and professional Dutch genealogist, Yvette Hoitink was gracious enough to provide an example of endogamy from an individual whose ancestors were from Winterswijk, a small village in the Netherlands.
Yvette tells us that in Winterswijk, people were serfs, some until 1795, and were required to pay a fine if they married a serf belonging to a different landlord.
Now, in addition to being a small village, we understand why so many people were related to each other, and why the other clusters are so tiny. Do note that many of the people in the red cluster also match people in the other colored clusters too, as identified by the grey cells. Truly, everyone does seem to be related to (at least) some of this person’s other ancestors.
Just so you don’t think all Dutch people are endogamous, Yvette also provided this autocluster of an individual from Friesland where people weren’t serfs during that timeframe.
Regional differences and population history, both on a large or small scale, really do make a HUGE difference.
Pedigree Collapse, Endogamy and Their Cousin, Population Genetics
I hope you have a better idea how pedigree collapse is different than endogamy and why endogamy is useful in population genetics.
- Pedigree collapse means you have the same ancestor(s) present in your tree, but other than those lines, it does NOT mean that everyone in your tree is related to each other.
- When everyone within a group is related somehow to everyone else, that’s endogamy.
- Of course, like many things in life, these “states of being” are not exclusive and entirely separate. You can have pedigree collapse without endogamy, but long-term community pedigree collapse within a group of people, such as the Acadians, defines endogamy.
- When endogamy is present, literally everyone is somehow related to everyone else, one way or another – especially distantly.
- You can have endogamy without any known recent ancestors.
- You can also have both pedigree collapse and endogamy, together, like my Acadian family line. If you do, I’m sorry😊!
With pedigree collapse, you have duplicate ancestors but you know who they are.
With endogamy, you’ll have a huge distant family, but it’s difficult or sometimes impossible to determine which ancestors, even if you DO know who they are, contributed specific DNA segments. Lots of matches with smaller matching DNA segments are prevalent and likely result from distant population-based matches.
_____________________________________________________________
Disclosure
I receive a small contribution when you click on some of the links to vendors in my articles. This does NOT increase the price you pay but helps me to keep the lights on and this informational blog free for everyone. Please click on the links in the articles or to the vendors below if you are purchasing products or DNA testing.
Thank you so much.
DNA Purchases and Free Transfers
- FamilyTreeDNA – Y, mitochondrial and autosomal DNA testing
- MyHeritage DNA – Autosomal DNA test
- MyHeritage FREE DNA file upload – Transfer your results from other vendors free
- AncestryDNA – Autosomal DNA test
- 23andMe Ancestry – Autosomal DNA only, no Health
- 23andMe Ancestry Plus Health
Genealogy Products and Services
- MyHeritage FREE Tree Builder – Genealogy software for your computer
- MyHeritage Subscription with Free Trial
- Legacy Family Tree Webinars – Genealogy and DNA classes, subscription-based, some free
- Legacy Family Tree Software – Genealogy software for your computer
- Charting Companion – Charts and Reports to use with your genealogy software or FamilySearch
- RootsMagic Software – Genealogy software for your computer
- Newspapers.com – Search newspapers for your ancestors
Books
- com – Lots of wonderful genealogy research books
Genealogy Research
- Legacy Tree Genealogists – Professional genealogy research
Discover more from DNAeXplained - Genetic Genealogy
Subscribe to get the latest posts sent to your email.













My mother’s autocluster report looks almost exactly like that Dutch example. Her biggest cluster is probably even 10% bigger. It was quite frustrating to see it first time. She was born in a small village in Finland.
Great article!
My ancestors were Ashkenazi Jews with some Sephardic origins. My maternal grandmother ‘s family tree shows pedigree collapse with several cousin marriages. For example, one of my 3rd cousins-once-removed matches me at the 2nd cousin level while her brother matches me at the 4th to 5th-cousin level.
I also share multiple tiny segments with non-Jewish NW Europeans and Latino Americans.
That’s very interesting. There were converses among the Spanish, but the NW Europeans is a bit of a mystery.
My parents were both born in Germany and, evidently both with distant Jewish ancestry (AJ and/or SJ). I am one of those non-Jewish NW Europeans matching AJ folks and Lation Americans. (One reason why I know that FTDNA has a sister company that markets to Brazilians. I see their kits uploaded at GEDMatch.) (I actually was born in Canada btw.)
Latino… I always type too fast. Sorry.
Thanks for a great article, Roberta. Nicely explained comparison. But are you missing a visual under the population genetics section or am I? I don’t see the Asian/Jewish comparison and the loss of colors/segments.
Hmm. I see it. If you’re looking at the email, check the website.
Nope, was looking at the website. But perhaps you were referring to the small segment endogamy figure way up in the endogamy paragraph? I was expecting a figure in the population genetics area.
Yes, that was the one. Maybe I should have located it further down.
I have noticed this with a 2nd cousin marriage where one of the grandparents was of African heritage. In addition, many of the kids from that 2nd cousin marriage married into one family. Most all married the brothers and sisters from another family unrelated to them who had no African ancestry. This seems to have lead some genealogists to incorrectly assume African origins attributed to the family they all married into and also to assume an African ancestor a generation later due to pedigree collapse (due to the 2nd cousin marriage). They see several cousins (many decedents) with same African DNA, and they then attribute this ancestor to the wrong side of the family not considering that all the brothers and sisters from one family marred into another and that the shared DNA came from from one family who married into the other family. Some looking for their roots are being led to believe the family my mixed race ancestors married into were their white slave owning ancestors, when the other side of the family simply happened to be free people of color and decedents from a second cousin marriage who married into this family on a large scale. These folks have (on their trees created with the assistance of a genealogist) my great-great grandmother as a mulatto sister of theirs and born into slavery not realizing that her husband’s parents “shared” grandfather was a free person of color and she was not a slave or even of African heritage. It was her husband’s family who had African heritage and many of his brothers and sisters also married her brothers and sisters. This is the source of my African DNA, as well. Thus, these folks are more likely related to a member of one of her husband’s other relatives who were also free people of color, some of which married into families of Native American, African American (some who may have been descended from slaves) or my case, of white origins, who lived in Louisiana, Mississippi, and the Carolinas.
Interesting diagram comparing the non-endogamous population with Ashkenazi and Native Americans. Could the amount of endogamy be used to get a handle on founding population size in Native Americans?
I don’t think so all this time later. But I will talk to a geneticist. It’s worth an ask at least. I love it when people think outside the box.
Thanks so much for a wonderful post. My husband is from St. Mary’s County, MD and his chart is really complicated. He has a number of ancestors where the the sons/daughters of one surname married the sons/daughters of another. He has one case where there were 3 sets. So all of those grandchildren only had one set of grandparents!
Best,
Winnie
My husband’s tree is complicated to say the least. He has lots of early VA folks. He has 3 brothers at the 7th Great Grandparent level, so of course that means the same 8th Great Grandparents are in his tree three times. At the 9th Great Grandparent level he has the same couple 4 times through the 3 brothers, and another line that connects to this couple. There are several other lines that he is connected through 2 different children of a couple, so descended from the same couple twice. He also has a line where brothers at the 7th Great Grandparents level married sisters, then there children married. The 6th Great Grandparents were double first cousins. There are so many repeated names in his tree. I know this is further back than you are talking about, but since there is so much of this in his tree would that cause his DNA matches to be *off*, and show higher than usual matches to people?
Yes, there certainly is. On the flip side, he’s more likely to have some of the DNA from those repeat ancestors further back in his tree.
Roberta, as usual your blog is interesting and informative. You tackled a difficult subject and made it easier to understand. My grandfather Edward is the son of Arthur. Edward married Anna Marie whose sister, Gretchen, married Arthurs father, Johan. Gretchen and Johan had two sons and a daughter. I keep thinking there is a double dose of DNA in there somewhere but I’m not sure where.
Hi Roberta, my mom’s German family is not a tree but a shrub, both her parents are related to each other and her maternal grandmother married her 1st cousin once removed (she married her grandfather’s brother’s son). RootsMagic calculates 12 different ways they are related. My mother’s parents are related from third cousin all the way to eight cousins. So I’m guessing I have both pedigree collapse and endogamy on my maternal side?
Just to note these were Germans who in late 1780 left Germany and settled in Bukovina/Bukowina, my family was primarily from Illischestie, and for 200+ years lived in the same village with very little marriage outside of religious or ethnic lines.
By the way loved you at RootsTech in 2020!
Thank you.
On my father’s (Winterswijk/endogamous) side, all my 2nd great-grandparents descend from the same couple, and share many other ancestors besides. Two of them lived the next village over but still shared ancestors with the rest, as you can see in the grey squares.
I had never heard of the authoritarian practice illustrated by your example of endogamy, Yvette! Very interesting. Did Dutch serfs keep track of their pedigree like higher social classes, in order to avoid getting riddled by hereditary diseases over time? This type of fine sounds like it carries long-term health risks, which the landlord must have known.
In my New York research, I encountered an article that explained that colonial Dutch families in and around the state of New York would have had written documentation of their roots, in other words, they would have known their family tree if they kept up with Dutch custom, at least for a few generations. However, was it common for all social classes to do this?
In French Canada, we kept track based on Roman Catholic canon law, and this ancestral practice triggers hilarity among my contacts from other ethnic groups… This musst be due to ignorance on their part, because obviously, Catholics couldn’t possibly have been the only Europeans aware of the importance of keeping track–and not just to be able to document one’s right to inherit.
So, I am curious about the ancestral documentation practices of European protestants of different social classes (in the lower classes of the Netherlands in this particular case).
Your moms cluster graph resembles my mom’s. One huge red block caused by pedigree collapse and endogomy. A series of interrelated families from VA Northern Neck that ended up in the Blue Ridge mtns.
I get confused when people talk also about bottlenecks/founder populations. If you scroll down this webpage you see the human population example of Dutch folks going to South Africa. https://evolution.berkeley.edu/evolibrary/article/bottlenecks_01 American Colonialists started off as a bottleneck population I guess. But after a few generations married outside their initial “gene pool”. This is not so much the case with the Ashkenazi Jewish folks I think. So much so, their DNA remains very distinctive to this day. Did I get this right???
Let me look at this when I get home. Writing about founder populations and bottlenecks would be a good idea too.
We tested my young great grandaughter whose mother is 1/2 Hawaiian from her mother’s side at MH. My ggrand’s top matches, and going down deep into the list, are from New Zealand and Australia with big amounts of cMs. I had not been able to square the large amounts of cMs, but your mentioning that new populations were not coming into their pool now makes sense of it. She has 7,000 matches. MH has a good representation of testers from that area. In just scrolling, she had a good match with a Fin. Do not know where that came from. Many thanks.
Excellent exposition of pedigree collapse in relation to DNA genealogy.
Of course, we all have pedigree collapse if it’s possible to go back far enough.
Transcripts of Visitations and some College of Arms records have one of my family lines back 32 generations to around 1100, when there are over 7 billion people in that generation in my tree. But there were not that many people in the world, let alone Britain, so some of those ancestors must be the same. One line apparently connects15 generations back.
The rest I have to take on trust due to the inevitability of the arithmetic.
And my preference for working much closer to the present day.
When pedigree collapse can have quite an effect on DNA calculations as you show.
Pingback: DNA: In Search Of…Your Grandparents | DNAeXplained – Genetic Genealogy
My parents are second cousins. Go back beyond 1830, and there are first cousin marriages and other odd permutations. I had a woman contact me, a second cousin, according to DNA, who was so excited, thinking she’d found the link to a long lost great grandfather. Through research I found our link in the 1700’s. Nothing like that old double dip of DNA.