Haplogroup D is a very old branch of Y-DNA that has remained rather mysterious. It has been uncertain where haplogroup D was born – in Africa, Asia or elsewhere – and when. It’s always fascinating when new research sheds light on the early history of humanity – discovered through people living and testing today.
In the current issue of Genetics, the article A Rare Deep-Rooting African Y-chromosomal Haplogroup and its Implications for the Expansion of Modern Humans Out of Africa by Haber et al appeared.
Their abstract:
Present-day humans outside Africa descend mainly from a single expansion out ∼50,000-70,000 years ago, but many details of this expansion remain unclear, including the history of the male-specific Y chromosome at this time. Here, we re-investigate a rare deep-rooting African Y-chromosomal lineage by sequencing the whole genomes of three Nigerian men described in 2003 as carrying haplogroup DE* Y-chromosomes, and analyzing them in the context of a calibrated worldwide Y-chromosomal phylogeny. We confirm that these three chromosomes do represent a deep-rooting DE lineage, branching close to the DE bifurcation, but place them on the D branch as an outgroup to all other known D chromosomes, and designate the new lineage D0. We consider three models for the expansion of Y lineages out of Africa ∼50,000-100,000 years ago, incorporating migration back to Africa where necessary to explain present-day Y-lineage distributions. Considering both the Y-chromosomal phylogenetic structure incorporating the D0 lineage, and published evidence for modern humans outside Africa, the most favored model involves an origin of the DE lineage within Africa with D0 and E remaining there, and migration out of the three lineages (C, D and FT) that now form the vast majority of non-African Y chromosomes. The exit took place 50,300-81,000 years ago (latest date for FT lineage expansion outside Africa – earliest date for the D/D0 lineage split inside Africa), and most likely 50,300-59,400 years ago (considering Neanderthal admixture).
Haplogroup DE was and is very rare. Because of its rarity, and that it had initially been reported in one man from Guinea-Bissau in West Africa and two Tibetans, it was unclear where DE originated, or when.
This new paper sequenced three men from Africa and five from Tibet.
D Splits
The result of the paper is that the authors confirm that the DE lineage split consists of three branches:
- E which is “mainly African” which we’ve known for a long time
- D0 which is exclusively African with the 3 Nigerian samples being within 2500 years of each other
- D which is exclusively non-African
To calibrate the branch length between any two samples when calculating split times, the authors multiplied the number of derived variants (mutations) found in the first sample but absent from the record, meaning previously unknown.
In supplementary table S2, they recalculate the splits between the various haplogroups. I found the table confusing to read, so I reached out to Goran Runfeldt who heads the scientific research team at Family Tree DNA to make this simpler.
I knew from previous discussions with the team that they had split the haplogroup D line internally to reflect a new branch at the time they named D-FT75 and subsequently D-FT76, and they were waiting for verification from multiple tests before splitting the line further.
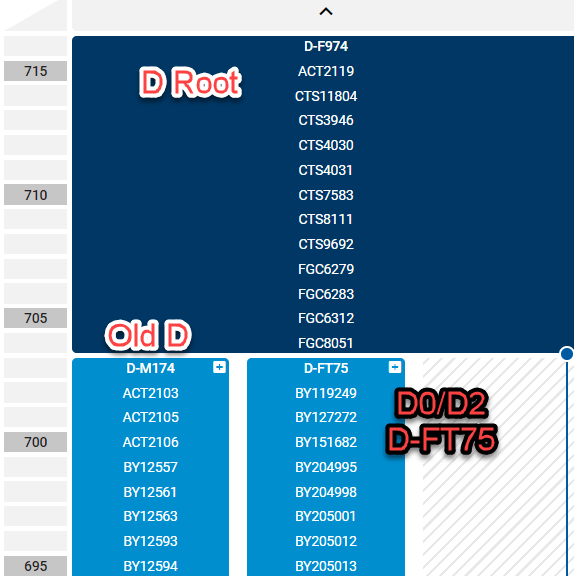
On the Family Tree DNA block tree, above, you can see the D split between D-F974 which is the main haplogroup D root (navy blue) which then splits into D-M174 which is the old line referred to as Haplogroup D, and the new D0/D2/D-FT75 lineage, both in lighter blue. You can see the public tree, here.
Goran explained that Family Tree DNA has actually found multiple lineages in what the authors call D0, which ISOGG calls D2 and Family Tree DNA refers to by the defining SNP as D-FT75.
If you’re like me, looking at this information in pedigree format is easier to comprehend.
I asked Goran and Big Y haplotree guru, Michael Sager if they could create something easy to understand. You can see them working together in this photo. Thanks guys!

The Haplogroup D Tree
Note that the following graphic is NOT TO TIME SCALE. Currently tested, unplaced and and pending samples are at the bottom.

In the chart above, haplogroups in red at the top are the base haplogroups, not refined by the paper. Green is the already known upper structure of haplogroup D. Tan is the haplogroup D structure being refined by Family Tree DNA. The blue group is the Nigerian structure from the paper.
Divergence times as quoted in the paper are noted. For example, the time between the split between CT and BT, according to the paper, is approximately 101.1 thousand years ago. (kya means thousands of years ago)
How the D-FT75 Branch was Discovered
At the end of 2018, Family Tree DNA published the first SNPs from the new haplogroup D lineage to the ISOGG SNP index. During 2019, additional SNPs have been added, including the new haplogroup D lines of D-FT75 and D-FT76.
I asked Michael Sager how he made that discovery.
When a customer purchases an STR test, if Family Tree DNA cannot reliably predict a haplogroup, they will run a backbone test, at no additional charge to the customer, to test enough SNPs to at least call a base level haplogroup, such as R-M269.
In this case, Family Tree DNA ran a backbone test on a customer’s Y DNA and the result came back as something Michael had never seen before – haplogroup CT, but no subgroup. As you’ve already noticed, haplogroup CT is far up the tree and Michael needed more information.
Michael said that he knew the only possible options were:
- CT* – where star means there is no subgroup. An individual with no CT subgroup has never been found, to date
- A lineage that breaks CT into a further haplogroup
- Haplogroup DE*
- A lineage that breaks haplogroup DE into further branches
- A lineage that breaks haplogroup D into further branches
- A lineage that breaks haplogroup E into further branches
After the backbone results were returned, Family Tree DNA contacted the customer and asked permission to run a Big Y test. The result was the discovery and naming of D-FT75 and D-FT76 which split D, twice, into new subgroups.
Further testing has verified the haplogroup D-FT76 finding in another Saudi Arabian male. Two additional haplogroup D males have results pending – one from Syria and one from another part of the world.
We now know that indeed the new branch of D, D0/D2/D-F75 has been found outside Africa, specifically in Saudi Arabia. It’s possible that there are more than two distinct lineages. We’ll know more as pending results come back from the lab.
However, what can be added is that according to the paper, the age of haplogroup D to the Nigerian samples is 71,400 years. The Family Tree DNA calculations based on the total number of 702 SNPs at 100 years per SNP suggest that the age is 70,200, which is very close to the 71,400 age in the paper. Additionally, because of the haplogroup FT75 and FT76 split, we can estimate the age of the divergence of those two lines with 261 SNPs between them at between 26,000 and 26,500 years, using these two calculation methods.
To quote Michael Sager, it’s “pretty neat to find a 20,000+-year-old NEW branch off of a 70,000+-year-old NEW branch.” I’d certainly agree!
Family Tree DNA would also like to place the Nigerian samples precisely on the tree.
In the supplemental data, the paper provided a list of the HG19 SNPs that are positive, including the positions for both D-FT75 and D-FT76, but did not list the SNPs that were negative. In order for Family Tree DNA to assign the Nigerian samples from the paper precisely to a branch, they need the BAM file because they need to see positive, negative and no-call SNPs. Family Tree DNA would also need to convert the results from build HG19, used by the authors, to current HG38.
What About You?
If you’re a male and have taken a Y STR test, meaning the 12, 25, 37, 67 or 111 marker test and you do not have a predicted haplogroup, please contact support at Family Tree DNA.
The best thing you can do, if you haven’t Y DNA tested, is to actually take a Y DNA test at Family Tree DNA. You can start out with the STR marker test which provides you with STR marker results, matching to other males and a haplogroup prediction.
Many individuals also purchase the Big Y-700 test which provides a very granular haplogroup – the most detailed possible, matching and at least 700 STR marker results – in addition to revealing never before discovered SNPs. Without the Big Y test, D-FT75 and D-FT76 and most of the 150,000 Y SNPs would not yet be discovered. This is the only test that can make new discoveries like this.
To summarize, you can be a part of scientific discovery if you’re a male (only males have Y chromosomes) by either:
- Testing your Y DNA by taking a 37, 67 or 111 marker test
- Ordering or upgrading to the Big Y-700 test
You can click here to order or upgrade.
______________________________________________________________
Disclosure
I receive a small contribution when you click on the link to one of the vendors in my articles. This does NOT increase the price you pay but helps me to keep the lights on and this informational blog free for everyone. Please click on the links in the articles or to the vendors below if you are purchasing products or DNA testing.
Thank you so much.
DNA Purchases and Free Transfers
- Family Tree DNA
- MyHeritage DNA only
- MyHeritage DNA plus Health
- MyHeritage FREE DNA file upload
- AncestryDNA
- 23andMe Ancestry
- 23andMe Ancestry Plus Health
- LivingDNA
Genealogy Services
Genealogy Research
- Legacy Tree Genealogists for genealogy research
Discover more from DNAeXplained - Genetic Genealogy
Subscribe to get the latest posts sent to your email.


My paternal 1st cousin, his dad, brother and uncle were given D/E haplogroup at 23andme years ago, they have not tested at FTDNA.
They sound like great candidates. Maybe you can share the article and they will test?
I mentioned it to him some time back to test with FTDNA, he is mildly interested, I would love for him to test. 23andme says their haplogroup is shared by 1 in 47,000 of their users, so there are others (some are from his paternal like). He is DE-M145,
From 23andme:
“Origin and Migrations of Haplogroup DE-M145
Haplogroup DE-M145 is so named because it traces back to the ancestral population of haplogroups D and E, as well as a few other rare lineages. The first man to bear the haplogroup likely lived between 65,000 and 76,000 years ago, but where he lived remains a mystery. Sometime between 50,000 and 70,000 years ago, a small group of people left Africa as part of the first major intercontinental migration by humans. It’s possible that men carrying the DE haplogroup journeyed from Africa across the Red Sea and into the Arabian peninsula. But it is also possible that DE-M145 originated within the Arabian Peninsula itself, and later spread back to Africa.
Within a few millennia the two groups of men in Africa and in Asia bearing the DE-M145 haplogroup became genetically distinct from each other. As a result, haplogroup D arose as a branch of DE-M145 in Asia, while haplogroup E arose as the African branch of DE-M145. Today the members of these two brother haplogroups are separated by thousands of miles: D is common in Japan, China and Tibet, whereas E is found in Africa, the Middle East, and southern Europe.
Yet the parent haplogroup of these two brothers has not disappeared, and persists even today. A few men in sub-Saharan Africa are linked by common origin to the DE-M145 haplogroup, but are not descendants of the D-M174 or E-M96 paternal lineages. Technically, those men still belong to the more ancient DE haplogroup, and are given the assignment DE-M145.”
Even if he purchases the STR test, it will give him a predicted haplogroup. If that’s interesting, then we can go from there. Where are his direct paternal ancestors from?
Probably Alabama, they are African-American
Hi Roberta, fantastic post. What’s the estimate for when the Saudi D2a samples split from the Nigerians? Also, has any analysis been done to compare any of these new samples to the Philippine D2?
We need to be able to place the Nigerian samples firmly on the tree and that can’t be done reliable without the BAM files. As for the D2, I don’t know. I’ll see what I can figure out next week.
It looks like ISOGG’s D2 is equivalent to Family Tree’s D.M226.2, both of which are nested under D-M174. So these new D2/D-FT75 samples are not in fact equivalent to ISOGG’s D2?
Here’s exactly what it says on the ISOGG page. “*D2 is listed as D0 in the 2019 Haber et al. study”
I will ask about a Phillipine D2 next week when FTDNA is open again, but no one mentioned it. Do you know if the Philippine D2 person tested there?
I’m not 100%, but I think kit YF03778 on YFull might be a D2 lineage (he’s currently just listed as a basal D*). The location is listed as Cebu in the Philippines. This is the only possible D2 kit I currently know of.
https://www.yfull.com/tree/D*/
I saw that too.
Two of the Filipino men in haplogroup D are the Doroy brothers; the father and uncle of my ex-girlfriend from Cebu (pronounced ceh-BOO, for those unfamiliar with this part of the world). Their kits which I administer are kits 233764 and N114828, respectively. Cebu is in the Visayas region where the people primarily speak Cebuano dialect. Far to the north is the capitol, Manila, where people primarily speak Tagalog dialect, and directly to the south is Mindinao, the island where Catholicism and Islam are present together, for the most part, peacefully.
There was also another Philippines haplogroup D kit N103885 with surname Pugay; I’m not sure what part of Philippines he is from. There was a predicted D kit from Guam with surname Macorol, of Chamorro ancestry, but the person administering the kit didn’t join the Haplogroup D Project and didn’t pursue any further testing, unfortunately. She did email me back and forth a few times, however, so it is possible someone else more assertive than me might be able to spark her interest in doing more advanced testing on her Chamorro kit. On (now-defunct) Ysearch, there was a kit listed as haplogroup DE from Ukraine, but the person didn’t respond when I sent them a message, and didn’t respond to a second email I sent them a few months later as a last desperate effort to make contact.
In 2012, curious about her ancestry, thinking that we might marry each other someday, and wanting to leave bread crumbs for our future children to find their distant relatives someday, I asked if I could test my penpal’s family members. She and her family granted permission. I ordered several FamilyFinder tests, and also Y tests for the males. When the Y results came back for her dad, I was dumbfounded. Predicted haplogroup D? In Philippines?! Sensing that his DNA could be imporant for haplogroup research, I ordered additional tests. The first series of additional tests were the SNP tests for each subclade of D. At the time, around 2013, there were three subclades of D. Her dad tested ancestral (negative) for every subclade, and he was thus placed in D* aka paragroup D or basal D. FamilyTreeDNA seemed to have trouble placing him and there were times when his kit was shown as predicted haplogroup C or S. He had no matches, not even at the 12 marker level, and seemed to be extremely distant from any other kit. On YHRD.org, he had zero matches for his haplotype across the entire worldwide database of about 40,000 kits. In Ysearch, he seemed to be closer to haplogroup E than to D, but with GD measurements extremely high (if I remember correctly, close to 100) when compared at 111 markers.
This was right around the time when Full Genomes was starting out. I had already ordered a Full Genomes kit for myself and was broke, so I took out a small loan from myself to order another kit for Mr. Doroy, thinking his results would be more important in the grand scheme of things. Several months later, in early 2014, the results were in. Full Genomes had discovered 579 (yes, 579!) new SNPs for Mr. Doroy. The new SNPs were FGC8841 to FGC9419. This was phenomenal. By comparison, mine in DF21 > L1336 “only” discovered 56 new SNPs, FGC11311 to FGC11366.
Fast forward a few months. My penpal was now my girlfriend. I wanted there to be a backup source of DNA in case her father’s DNA ran out, so I asked to test her uncle (her father’s brother). This would also confirm her father’s strange results. I ordered National Geographics Geno 2.0 for her uncle, transferred the results into FamilyTreeDNA, and immediately upgraded to Big Y, which “rediscovered” FGC8841 to FGC9419.
Mr. Pugay’s kit was instrumental in defining the new subclade. Comparing Pugay against Doroy, there were several dozen SNPs in common. The new subclade was initially named D-M226, but M226 had already been found in haplogroup M, so FamilyTreeDNA suffixed .2 and renamed the subclade D-M226.2. ISOGG’s name for the new subclade was D-L1366 until 2016, at which point L1366 was removed for reasons unknown, in favor of D-L1378. Defining the new subclade with its various names had the effect of turning the rest of haplogroup D upside-down. What had previously been called D1, D2, and D3 were now subsumed into the new D1, and the Philippines D-M226.2 aka D-L1378 became the new D2.
In 2019, the DE* research turned the D tree upside-down yet again, with D1 (Japan, China, Tibet) becoming the new D1a, D2 (Philippines) becoming D1b, and the Nigerian, Saudi Arabian, and Syrian kits becoming the new D2. Just a few weeks ago, I replied to an email from FamilyTreeDNA in which the company asked me for permission to upgrade Philippines kit N114828, and another unrelated kit from Papau New Guinea belonging to haplogroup M, to Big Y 700 for free. I called in as the email requested, and asked who wanted to do the free upgrade on my behalf. The customer service representative put me on hold for a few seconds and when she came back, told me that it was a researcher working in FamilyTreeDNA who is fascinated with haplogroup research into rare haplogroups. I said YES, permission granted! She then asked me to stay on hold for a few minutes. She came back and told me that “she was so happy and wanted me to express her great appreciation” or something to that effect. I suspect there is a female scientist doing research, comparing the Philippines kits against the Nigerian, Saudi Arabian, and Syrian kits.
Exciting!!
The Nigerian branch of D2a is very recent, its tmrca is 2.5ky, it can not be such a decisive argument.
Where are they from? What SNPs did they test?
What are the Philippine D2 ?
I don’t know, and I don’t know where they tested.
There are two phillipino D2(now D1b) in the ftDNA haplogroup D project. There used to be a third one but I can’t see it no more.
Thank you, Roberta!
Very interesting! Thanks once again for bringing this information to us, and for going to the trouble of making it more intelligible to the layperson.
Still, I am struggling to make sure I understand this. Like some others in these comments, I find different names being used for the same branches by different groups confusing. It must be challenging to add an earlier branching in an already extensive pedigree/tree, which is labeled alphabetically in chronological order – mostly, but not entirely – and which still has multiple naming conventions in use despite ISOGG’s efforts to standardize it. How do you name a newly discovered branch without creating more confusion?
I think they are saying that D2=D0. Then there’s D-FT75 and D-FT76, D.M226.2, and D-M174. Sounds like the jury’s out on exactly how these fit in until FTDNA has the information to fully analyze the results in the study. All you can do is relate the study’s assertion that D2=D0.
This is forcing me to improve my overall understanding of yDNA testing. I guess I need to remember that D0/D2 is an intellectual reconstruction of an ancient yDNA configuration, based on what persists today. You are saying that the Nigerian, Saudi Arabian and Syrian men mentioned are descended from D0/D2, not that they have D0/D2 exactly as it was 26.5 kya.
It looks to me like the Syrian and Saudi Arabian samples branched off earlier than the Nigerians according to this pedigree/tree – am I interpreting this correctly?
I am wondering about the Tibetans you mentioned – they are not shown in this tree. Is this an issue of needing more detailed testing? I’m curious how their results would compare to the Philipino D2 results, and what this would say about ancient migrations in Asia.
Roberta, Great article ! Can we have as a topic about how ONE hundred years is the period of time on average between single nucleotide polymorphism (SNP) mutations ?
With the increased number of BigY tests I suggest that the time period between mutations will be revised downwards. I think the data is showing a higher rate of mutation. Is it, that with next generation sequencing (NGS) the newer SNPs being tested are less stable than the SNPs used to previously.
Could we be able to use a good reliable pedigree ( say 15 generations ) allowing thirty years per generation and compare the triangulation between the males who can be placed in this pedigree; thereby giving a timeline to their common SNPs back to a common ancestor’s SNP
Tim
I found it interesting that the 100 years per SNP and the authors method arrived at very close to the same year.
Roberta, regarding this comment in your post: “Further testing has verified the haplogroup D-FT76 finding in another Saudi Arabian male. Two additional haplogroup D males have results pending – one from Syria and one from another part of the world.”
Do you know where the D sample “from another part of the world” is from specifically?
One thing I find interesting, on ISOGG’s D page it mentions that undifferentiated D* samples have been recorded in various studies. It would be great if some of these samples could eventually be located and sequenced – will they prove be a diverged branch within the newly christened D1, D2, or an entire new branch of D altogether?
We don’t have answers yet. I will write more when more information is available.
The TMRCA of the Nigerian branch is only 2.5 ky , the conclusion is unreliable
It’s not only that; the study (pages 4–5) finds that haplogroups DE, E, and E all diverged between 80,000 and 71,000 bc, and also cites the evidence that the Out-of-Africa migration to the Near East of the ancestors of modern Eurasians/non-Africans (and the neanderthal admixture that happened soon after) occurred significantly after (between about 50,000–60,000 years ago). This also (as the paper explains) indicates that DE, E, and D0 diverged in Africa before the Out of Africa migration (that is, before the Out-of-Africa migration of ca. 50,000 bc–60,000 bc that was ancestral to all modern non-Africans). (There was at least one earlier migration of modern humans Out of Africa around 120,000 bc, but it became mostly extinct and was replaced by the later ca 50-60,000 bc wave, surviving only in traces in just a few modern groups and surviving not at all in others—about 2% of the ancestry of Papua New Guineans and Australian Aboriginals comes from the earlier migration, but other modern non-African populations, as far as we know, have no “early migration” ancestry.
https://www.genetics.org/content/genetics/early/2019/06/13/genetics.119.302368.full.pdf
“All non-Africans carry around 2% Neanderthal DNA in their genomes (Green et al. 2010), and Neanderthal fossils have only been reported outside Africa. The geographical distribution of Neanderthals thus suggests that the mixing probably occurred outside Africa, and the ubiquitous presence of Neanderthal DNA in present-day non-Africans is most easily explained if the mixing took place once, soon after the migration out. This mixing has been dated with some precision using the length of the introgressed segments in the 45,000-year-old (43,210- 46,880 years) Siberian (Ust’-Ishim) to 232-430 generations before he lived, i.e. 49,900-59,400 years ago assuming a generation time of 29 years (Fu et al. 2014). If this date represented the time of the migration out of Africa, it would exclude the first two scenarios (Figure 2B and 2C) [2B and 2C being the scenarios involving a Eurasian back-migration of E and D0, and 2D being the third and more likely scenario involving an African origin of E and D0 as well as DE]. Thus the combination of Y phylogenetic structure and dating of the out-of-Africa migration based on the 45,000-year-old Siberian fossil (Fu et al. 2014) favors the third scenario (Figure 2D) involving the migration out of C, D and FT between 50,300 years ago (lower bound of the FT diversification, Table S2) and 59,400 years ago (upper bound of the introgression; see Figure 3) which is in accordance with suggested models incorporating an African origin of the DE lineages…”
It seems that a plausible scenario may be that: DE may have originated in Africa as the paper suggests, seemingly likely East Africa (with E also originating/diverging there and mostly staying in Africa—with one branch, E1b1b, later leaving for the Near East). And the D/D0 split could also have happened in (or near) East Africa, with D going east to Asia, and with some D0 moving deeper into Africa eventually to West Africa (e.g. Nigeria) and other D0 moving toward the Middle East (and with D0, as far as we know, dying out in regions in-between—leaving a few derived remnant lineages in both Africa and the Near East)—It seems that its (D0’s) origin might likely have been in between the regions where it is now found (i.e. possibly originating in East Africa/northeast Africa, in between West Africa and the Middle East/West Asia—it is now found deep in Africa on the one hand, and in the Middle East near Africa on the other).
It also seems possible that the Middle Eastern D0, or some of the Middle Eastern D0 (especially the Saudi Arabian) could possibly be the result of African gene flow into the Near East in more recent times (as was sometimes suggested for the proposed DE there—before it was reclassified as D0), the Arab slave trade being one episode of this, and there is evidence of small amounts of African gene flow in much of the Middle East (some of it ultimately deriving from West/Central Africa specifically). Some of the slaves brought to the Middle East came from the Bantu ethnic groups of Eastern Africa (Kenya, Tanzania, etc) who had ancestry ultimately originating from the Bantu homeland around Cameroon, not far from southeast Nigeria where the Nigerian D0 is found (which is, I believe, the Cross River region, not far from the Bantu homeland in/around Cameroon). D0 is, like DE, now very rare today both in Africa and Asia (likely once being more widespread but having largely died out/been mostly replaced by other haplogroups).
But it would be great to find ancient y-dna from the relevant periods in various parts of Africa and Eurasia, preferably mesolithic or earlier (however likely that may be). Hopefully, at the least, the purported Tibetan “DE”, which the authors of the paper (Haber et al.) suggest may perhaps not be true DE* (but possibly a back-mutation or lab error) and suggest should be re-analyzed, will be analyzed soon (along with the interesting newly discovered basal Filipino D lineages).
(Minor clarification): …the study by Haber et al (pages 1425–1426 of pdf, and Table S2 of Supplemental Material) finds that haplogroups DE, E, and D0 all likely diverged between about 76,000 and 71,000 years ago, and also finds that the Out-of-Africa migration of the ancestors of modern Eurasians/non-Africans (and the neanderthal admixture that happened soon after) most likely occurred significantly later than that (between about 50,000–60,000 years ago). This is one of the reasons (as the paper explains) that DE, E, and D0 are considered likely by the authors to have originated in Africa (i.e. somewhere within Africa before the OOA migration of ancestral Eurasians).
(Note: I earlier accidentally posted this comment with an error. This is the corrected version. My apologies. I did not mean to double post)
It’s not only that; the study (pages 4–5) finds that haplogroups DE, E, and D0 all diverged between 80,000 and 71,000 bc, and also cites the evidence that the Out-of-Africa migration to the Near East of the ancestors of modern Eurasians/non-Africans (and the neanderthal admixture that happened soon after) occurred significantly after (between about 50,000–60,000 years ago). This also (as the paper explains) indicates that DE, E, and D0 diverged in Africa before the Out of Africa migration (that is, before the Out-of-Africa migration of ca. 50,000 bc–60,000 bc that was ancestral to all modern non-Africans). (There was at least one earlier migration of modern humans Out of Africa around 120,000 bc, but it became mostly extinct and was replaced by the later ca 50-60,000 bc wave, surviving only in traces in just a few modern groups and surviving not at all in others—about 2% of the ancestry of Papua New Guineans and Australian Aboriginals comes from the earlier migration, but other modern non-African populations, as far as we know, have no “early migration” ancestry.
https://www.genetics.org/content/genetics/early/2019/06/13/genetics.119.302368.full.pdf
“All non-Africans carry around 2% Neanderthal DNA in their genomes (Green et al. 2010), and Neanderthal fossils have only been reported outside Africa. The geographical distribution of Neanderthals thus suggests that the mixing probably occurred outside Africa, and the ubiquitous presence of Neanderthal DNA in present-day non-Africans is most easily explained if the mixing took place once, soon after the migration out. This mixing has been dated with some precision using the length of the introgressed segments in the 45,000-year-old (43,210- 46,880 years) Siberian (Ust’-Ishim) to 232-430 generations before he lived, i.e. 49,900-59,400 years ago assuming a generation time of 29 years (Fu et al. 2014). If this date represented the time of the migration out of Africa, it would exclude the first two scenarios (Figure 2B and 2C) [2B and 2C being the scenarios involving a Eurasian back-migration of E and D0, and 2D being the third and more likely scenario involving an African origin of E and D0 as well as DE]. Thus the combination of Y phylogenetic structure and dating of the out-of-Africa migration based on the 45,000-year-old Siberian fossil (Fu et al. 2014) favors the third scenario (Figure 2D) involving the migration out of C, D and FT between 50,300 years ago (lower bound of the FT diversification, Table S2) and 59,400 years ago (upper bound of the introgression; see Figure 3) which is in accordance with suggested models incorporating an African origin of the DE lineages…”
It seems that a plausible scenario may be that: DE may have originated in Africa as the paper suggests, seemingly likely East Africa (with E also originating/diverging there and mostly staying in Africa—with one branch, E1b1b, later leaving for the Near East). And the D/D0 split could also have happened in (or near) East Africa, with D going east to Asia, and with some D0 moving deeper into Africa eventually to West Africa (e.g. Nigeria) and other D0 moving toward the Middle East (and with D0, as far as we know, dying out in regions in-between—leaving a few derived remnant lineages in both Africa and the Near East)—It seems that its (D0’s) origin might likely have been in between the regions where it is now found (i.e. possibly originating in East Africa/northeast Africa, in between West Africa and the Middle East/West Asia—it is now found deep in Africa on the one hand, and in the Middle East near Africa on the other).
It also seems possible that the Middle Eastern D0, or some of the Middle Eastern D0 (especially the Saudi Arabian) could possibly be the result of African gene flow into the Near East in more recent times (as was sometimes suggested for the proposed DE there—before it was reclassified as D0), the Arab slave trade being one episode of this, and there is evidence of small amounts of African gene flow in much of the Middle East (some of it ultimately deriving from West/Central Africa specifically). Some of the slaves brought to the Middle East came from the Bantu ethnic groups of Eastern Africa (Kenya, Tanzania, etc) who had ancestry ultimately originating from the Bantu homeland around Cameroon, not far from southeast Nigeria where the Nigerian D0 is found (which is, I believe, the Cross River region, not far from the Bantu homeland in/around Cameroon). D0 is, like DE, now very rare today both in Africa and Asia (likely once being more widespread but having largely died out/been mostly replaced by other haplogroups).
But it would be great to find ancient y-dna from the relevant periods in various parts of Africa and Eurasia, preferably mesolithic or earlier (however likely that may be). Hopefully, at the least, the purported Tibetan “DE”, which the authors of the paper (Haber et al.) suggest may perhaps not be true DE* (but possibly a back-mutation or lab error) and suggest should be re-analyzed, will be analyzed soon (along with the interesting newly discovered basal Filipino D lineages).
Edit:
“…DE, E, and D0 all most likely diverged between 80,000 and 71,000 bc,—or (as the tree shows) more precisely, divergence dates of: 67,600-86,700 years ago for DE (most likely about 76,500 years ago), 64,700-83,000 years ago for D0/D and E (most likely about 73,000 years ago), and 63,000-83,000 years ago (most likely ca, 71,000 years ago) for D0,…”
(the migration out of Africa for the ancestors of modern non-Africans being estimated at about. 49,900-59,400 years ago)
(The quoted section/passage in my first comment above is, of course, from the paper.)
Thanks for this article Roberta.
Do you know if FTDNA funds tests for “rare”/under-tested branches of the haplotree?
We’re particularly interested in supporting the development of C-M38. We’ve paid for two Big Y recently and have other kits we could add Big Y on to but can’t afford the upgrades at this stage.
Any suggestions for funding are appreciated.
Thanks, Elena
D-M174 here, as well as A00, J-M172, J-M410, and I’m a Kohanim, and it’s much more). And I would be considered African American ). Crazy times huh ?