Are you confused about DNA matches and what they mean…different kinds of matches…from different vendors and combined results between vendors. Do you feel like lions and tigers and bears…oh my? You’re not alone.
As the vendors add more tools, I’ve noticed recently that along with those tools has come a significant amount of confusion surrounding matches and what they mean. Add to this issue confusion about the terminology being used within the industry to describe various kinds of matches. Combined, we now have a verbiage or terminology issue and we have confusion regarding the actual matches and what they mean. So, as people talk, what they mean, what they are trying to communicate and what they do say can be interpreted quite widely. Is it any wonder so many people are confused?
I reached out within the community to others who I know are working with autosomal results on a daily basis and often engaged in pioneering research to see how they are categorizing these results and how they are referring to them.
I want to thank Jim Bartlett, Blaine Bettinger, Tim Janzen and David Pike (in surname alphabetical order) for their input and discussion about these topics. I hope that this article goes a long way towards sorting through the various kinds of matches and what they can and do mean to genetic genealogists – and what they are being called. To be clear, the article is mine and I have quoted them specifically when applicable.
But first, let’s talk about goals.
Goals
One thing that has become apparent over the past few months is that your goals may well affect how you interpret data. For example, if you are an adoptee, you’re going to be looking first at your closest matches and your largest segments. Distant matches and small segments are irrelevant at least until you work with the big pieces. The theory of low hanging fruit, of course.
If your goal is to verify and generally validate your existing genealogy, you may be perfectly happy with Ancestry’s Circles. Ancestry Circles aren’t proof, as many people think, but if you’re looking for low hanging fruit and “probably” versus “positively,” Ancestry Circles may be the answer for you.
If you didn’t stop reading after the last sentence, then I’m guessing that “probably” isn’t your style.
If your goal is to prove each ancestor and/or map their segments to your DNA, you’re not going to be at all happy with Ancestry’s lack of segment data – so your confidence and happiness level is going to be greatly different than someone who is just looking to find themselves in circles with other descendants of the same ancestor and go merrily on their way.
If you have already connected the dots on most of your ancestry for the past 4 or 5 generations, and you’re working primarily with colonial ancestors and those born before 1700, you may be profoundly interested in small segment data, while someone else decides to eliminate that same data on their spreadsheet to eliminate clutter. One person’s clutter is another’s goldmine.
While, technically, the different types of tests and matches carry a different technical confidence level, your personal confidence ranking will be influenced by your own goals and by some secondary factors like how many other people match on a particular segment.
Let’s start by talking about the different kinds of matching. I’ve been working with my Crumley line, so I’ll be utilizing examples from that project.
Individual Matching, Group Matching and Triangulation
There is a difference between individual matching, group matching and triangulation. In fact, there is a whole spectrum of matching to be considered.
Individual Matching
Individual matching is when someone matches you.
That’s great, but one match out of context generally isn’t worth much. There’s that word, generally, because if there is one thing that is almost always true, it’s that there is an exception to every rule and that exception often has to do with context. For example, if you’re looking for parents and siblings, then one match is all you need.
If this match happens to be to my first cousin, that alone confirms several things for me, assuming there is not a secondary relationship. First, it confirms my relationship with my parent and my parent’s descent from their parents, since I couldn’t be matching my first cousin (at first cousin level) if all of the lines between me and the cousin weren’t intact.
However, if the match is to someone I don’t know, and it’s not a close relative, like the 2nd to 4th cousins shown in the match above, then it’s meaningless without additional information. Most of your matches will be more distant. Let’s face it, you have a lot more distant cousins than close cousins. Many ancestors, especially before about 1900, were indeed, prolific, at least by today’s standards.
So, at this point, your match list looks like this:
Bridget looks pretty lonely. Let’s see what we can do about that.
Matching Additional People
The first question is “do you share a common ancestor with that individual?” If yes, then that is a really big hint – but it’s not proof of anything – unless they are a close relative match like we discussed above.
Why isn’t a single match enough for proof?
You could be related to this person through more than one ancestral line – and that happens far more than I initially thought. I did an analysis some time back and discovered that about 15% of the time, I can confirm a secondary genealogical line that is not related to the first line in my tree. There were another 7% that were probable – meaning that I can’t identify a second common ancestor with certainty, but the surname and location is the same and a connection is likely. Another 8% were from endogamous lines, like Acadians, so I’m sure there are multiple lines involved. And of those matches (minus the Acadians), about 10% look to have 3 genealogical lines, not just two. The message here – never assume.
When you find one match and identify one common genealogical line, you can’t assume that is how you are genetically related on the segment in question.
Ideally, at this point, you will find a third person who shares the common ancestor and their DNA matches, or triangulates, between you and your original match to prove the connection. But, circumstances are not always ideal.
What is Triangualtion?
Triangulation on the continuum of confidence is the highest confidence level achievable, outside of close relative matching which is evident by itself without triangulation.
Triangulation is when you match two people who share a common ancestor and all three of you match each other on that same segment. This means that segment descended to all three of you from that common ancestor.
This is what a match group would look like if Jerry matches both John and Bridget.
Example 1 – Match Group
The classic definition of triangulation is when three people, A, B and C all match each other on the same segment and share a known, identifiable common ancestor. Above, we only have two. We don’t know yet if John matches Bridget.
A matches B
A matches C
B matches C
This is what an exact triangulation group would look like between Jerry, John and Bridget. Most triangulation matches aren’t exact, meaning the start and/or end segment might be different, but some are exact.
Example 2 – Triangulation Group
It’s not always possible to prove all three. Sometimes you can see that Jerry matches Bridget and Jerry matches John, but you have no access to John or Bridget’s kits to verify that they also match each other. If you are at Family Tree DNA, you can run the ICW (in common with) tool to see if John and Bridget do match each other – but that tool does not confirm that they match on the same segment.
If the individuals involved have uploaded their kits to GedMatch, you have the ability to triangulate because you can see the kit numbers of your matches and you can then run them against each other to verify that they do indeed match each other as well. Not everyone uploads their kits to GedMatch, so you may wind up with a hybrid combination of triangulated groups (like example 2, above) and matching groups (like example 1, above) on your own personal spreadsheet.
Matching groups (that are not triangulated) are referred to by different names within the community. Tim Janzen refers to them as clusters of cousins, Blaine as pseudo triangulation and I have called them triangulation groups in the past if any three within the group are proven to be triangulated. Be careful when you’re discussing this, because matching groups are often misstated as triangulated groups. You’ll want to clarify.
Creating a Match List
Sometimes triangulation options aren’t available to us. For example, at Family Tree DNA, we can see who matches us, and we can see if they match each other utilizing the ICW tool, but we can’t see specifically where they match each other. This is considered a match group. This type of matching is also where a great deal of confusion is introduced because these people do match each other, but they are NOT (yet) triangulated.
What we know is that all of these people are on YOUR match list, but we don’t know that they are on each other’s match lists. They could be matching you on different sides of your DNA or, if smaller segments, they might be IBC (identical by chance.)
You can run the ICW (in common with) tool at Family Tree DNA for every match you have. The ICW tool is a good way to see who matches both people in question. Hopefully, some of your matches will have uploaded trees and you can peruse for common ancestors.
The ICW tool is the little crossed arrows and it shows you who you and that person also match in common.
You can run the ICW tool in conjunction with the ancestral surname in question, showing only individuals who you have matches in common with who have the Crumley surname (for example) in their ancestral surname list. This is a huge timesaver and narrows your scope of search immediately. By clicking on the ICW tool for Ms. Bridget, you see the list, below of those who match both the person whose account we are signed into and Ms. Bridget, below.
Another way to find common matches to any individual is to search by either the current surname or ancestral surnames. The ancestral surname search checks the surnames entered by other participants and shows them in the results box.
In the example above, all of these individuals have Crumley listed in their surnames. You can see that I’ve sorted by ancestral surname – as Crumley is in that search box.
Now, your match lists looks like this relative to the Crumley line. Some people included trees and you can find your common ancestor on their tree, or through communications with them directly. In other cases, no tree but the common surname appears in the surname match list. You may want to note those results on your match list as well.
Of course, the next step is to compare these individuals in a matrix to see who matches who and the chromosome browser to see where they match you, which we’ll discuss momentarily.
Group Matching
The next type of matching is when you have a group of people who match each other, but not necessarily on the same segment of DNA. These matching groups are very important, especially when you know there is a shared ancestor involved – but they don’t indicate that the people share the same segment, nor that all (or any) of their shared segments are from this particular ancestor. Triangulation is the only thing that accomplishes proof positive.
This ICW matrix shows some of the Crumley participants who have tested and who matches whom.
You can display this grid by matching total cM or by known relationship (assuming the individuals have entered this information) or by predicted relationship range. The total cMs shared is more important for me in evaluating how closely this person might be related to the other individual.
The Chromosome Browser
The chromosome browser at Family Tree DNA shows matches from the perspective of any one individual. This means that the background display of the 22 Chromosomes (plus X) is the person all of the matches are comparing against. If you’re signed in to your account, then you are the black background chromosomes, and everyone is being compared against your DNA. I’m only showing the first 6 chromosomes below.
You can see where up to 5 individuals match the person you’re comparing them to. In this case, it looks like they may share a common segment on chromosome 2 among several descendants. Of course, you’d need to check each of these individuals to insure that they match each other on this same segment to confirm that indeed, it did come from a common ancestor. That’s triangulation.
When you see a grouping of matches of individuals known to descend from a common ancestor on the same chromosome, it’s very likely that you have a match group (cluster of cousins, pseudo triangulation group) and they will all match each other on that same segment if you have the opportunity to triangulate them, but it’s not absolute.
For example, below we have a reconstructed chromosome 8 of James Crumley, the common ancestor of a large group of people shown based on matches. In other words, each colored segment represents a match between two people. I have a lot more confidence in the matches shown with the arrows than the single or less frequent matches.
This pseudo triangulation is really very important, because it’s not just a match, and it’s not triangulation. The more people you have that match you on this segment and that have the same ancestor, the more likely that this segment will triangulate. This is also where much of the confusion is coming from, because matching groups of multiple descendants on the same segments almost always do triangulate so they have been being called triangulation groups, even when they have not all been triangulated to each other. Very occasionally, you will find a group of several people with a common ancestor who triangulate to each other on this common segment, except one of a group doesn’t triangulate to one other, but otherwise, they all triangulate to others.
This situation has to be an error of some sort, because if all of these people match each other, including B, then B really must match D. Our group discussed this, and Jim Bartlett pointed out that these problem matches are often near the vendor matching threshold (or your threshold if you’re using GedMatch) and if the threshold is lowered a bit, they continue to match. They may also be a marginal match on the edge, so to speak or they may have a read error at a critical location in their kit.
What “in common with” matching does is to increase your confidence that these are indeed ancestral matches, a cousin cluster, but it’s not yet triangulation.
Ancestry Matches
Ancestry has added another level of matching into the mix. The difference is, of course, that you can’t see any segment data at all, at Ancestry, so you don’t have anything other than the fact that you do match the other person and if you have a shakey leaf hint, you also share a common ancestor in your trees.
When three people match each other on any segment (meaning this does not infer a common segment match) and also share a common ancestor in a tree, they qualify to be a DNA Circle. However, there is other criteria that is weighted and not every group of 3 individuals who match and share an ancestor becomes a DNA Circle. However, many do and many Circles have significantly more than three individuals.
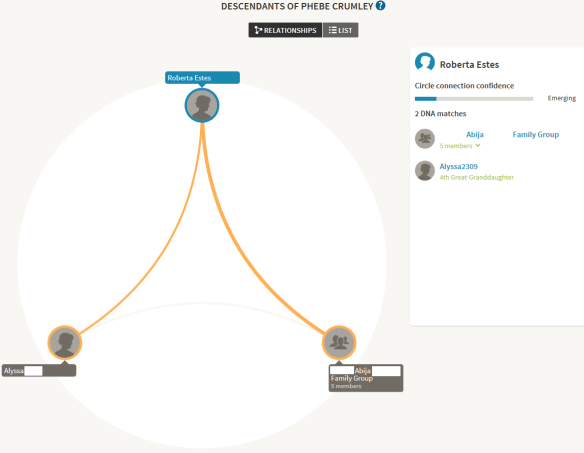
This DNA Circle is for Phebe Crumley, one of my Crumley ancestors. In this grouping, I match one close family group of 5 people, and one individual, Alyssa, all of whom share Phebe Crumley in their trees. As luck would have it, the family group has also tested at Family Tree DNA and has downloaded their results to GedMatch, but as it stands here at Ancestry, with DNA Circle data only…the only thing I can do is to add them to my match list.
In case you’re wondering, the reason I only added three of the 5 family members of the Abija group to my match list is because two are children of one of the members and their Crumley DNA is represented through their parent.
While a small DNA Circle like Phebe Crumley’s can be incorrect, because the individuals can indeed be sharing the DNA of a different ancestor, a larger group gives you more confidence that the relationship to that group of people is actually through the common ancestor whose circle you are a member of. In the example Circle shown below, I match 6 individuals out of a total of 21 individuals who are all interrelated and share Henry Bolton in their tree.
New Ancestor Discoveries
Ancestry introduced New Ancestor Discoveries (NADs) a few months ago. This tool is, unfortunately, misnamed – and although this is a good concept for finding people whose DNA you share, but whose tree you don’t – it’s not mature yet.
The name causes people to misinterpret the “ancestors” given to them as genuinely theirs. So far, I’ve had a total of 11 NADS and most have been easily proven false.
Here’s how NADs work. Let’s say there is a DNA Circle, John Doe, of 3 people and you match two of them. The assumption is that John Doe is also your ancestor because you share the DNA of his descendants. This is a critically flawed assumption. For example, in one case, my ancestors sister’s husband is shown as my “new ancestor discovery” because I share DNA with his descendants (through his wife, my ancestor’s sister.) Like I said, not mature yet.
I have discussed this repeatedly, so let’s just suffice it to say for this discussion, that there is absolutely no confidence in NADs and they aren’t relevant.
Shared Matches
Ancestry recently added a Shared Matches function.
For each person that you match at Ancestry, that is a 4th cousin or closer and who has a high confidence match ranking, you can click on shared matches to see who you and they both match in common.
This does NOT mean you match these people through the same ancestor. This does NOT mean you match them on the same segment. I wrote about how I’ve used this tool, but without additional data, like segment data, you can’t do much more with this.
What I have done is to build a grid similar to the Family Tree DNA matrix where I’ve attempted to see who matches whom and if there is someone(s) within that group that I can identify as specifically descending from the same ancestor. This is, unfortunately, extremely high maintenance for a very low return. I might add someone to my match list if they matched a group (or circle) or people that match me, whose common ancestor I can clearly identify.
Shared Matches are the lowest item on the confidence chart – which is not to say they are useless. They can provide hints that you can follow up on with more precise tools.
Let’s move to the highest confidence tool, triangulation groups.
Triangulation Groups
Of course, the next step, either at 23andMe, Family Tree DNA, through GedMatch, or some combination of each, is to compare the actual segments of the individuals involved. This means, especially at Ancestry where you have no tools, that you need to develop a successful begging technique to convince your matches to download their data to GedMatch or Family Tree DNA, or both. Most people don’t, but some will and that may be the someone you need.
You have three triangulation options:
- If you are working with the Family Inheritance Advanced at 23andMe, you can compare each of your matches with each other. I would still invite my matches to download to GedMatch so you can compare them with people who did not test at 23andMe.
- If you are working with a group of people at Family Tree DNA, you can ask them to run themselves against each other to see if they also match on the same segment that they both match you on. If you are a project administrator on a project where they are all members, you can do this cross-check matching yourself. You can also ask them to download their results to GedMatch.
- If your matches will download their results to GedMatch, you can run each individual against any other individual to confirm their common segment matches with you and with each other.
In reality, you will likely wind up with a mixture of matches on your match list and not everyone will upload to GedMatch.
Confirming that segments create a three way match when you share a common ancestor constitutes proof that you share that common ancestor and that particular DNA has been passed down from that ancestor to you.
I’ve built this confidence table relative to matches first found at Family Tree DNA, adding matches from Ancestry and following them to GedMatch. Fortunately, the Abija group has tested at all 3 companies and also uploaded their results to GedMatch. Some of my favorite cousins!
Spectrum of Confidence
Blaine Bettinger built this slide that sums up the tools and where they fall on the confidence range alone, without considerations of your goals and technical factors such as segment size. Thanks Blaine for allowing me to share it here.
These tools and techniques fall onto a spectrum of confidence, which I’ve tried to put into perspective, below.
I really debated how to best show these. Unfortunately, there is almost always some level of judgment involved. In some cases, like triangulation at the 3 vendors, the highest level is equivalent, but in other cases, like the medium range, it really is a spectrum from lowest to highest within that grouping.
Now, let’s take a look at our matches that we’ve added to our match list in confidence order.
As you would expect, those who triangulated with each other using some chromosome browser and share a common ancestor are the highest confidence matches – those 5 with a red Y. These are followed by matches who match me and each other but not on the same segment (or at least we don’t know that), so they don’t triangulate, at least not yet.
I didn’t include any low confidence matches in this table, but of the lowest ones that are included, the shakey leaf matches at Ancestry that won’t answer inquiries and the matches at FTDNA who do share a common surname but didn’t download their information to be triangulated are the least confident of the group. However, even those lower confidence matches on this chart are medium, meaning at Ancestry they are in a Circle and at FTDNA, they do match and share a common surname. At Family Tree DNA, they may eventually fall into a triangulation group of other descendants who triangulate.
Caveats
As always, there are some gotchas. As someone said in something I read recently, “autosomal DNA is messy.”
Endogamy
Endogamous populations are just a mess. The problem is that literally, everyone is related to everyone, because the founder population DNA has just been passed around and around for generations with little or no new DNA being introduced.
Therefore, people who descend from endogamous populations often show to be much more closely related than they are in a genealogical timeframe.
Secondly, we have the issue pointed out by David Pike, and that is when you really don’t know where a particular segment came from, because the segment matches both the parents, or in some cases, multiple grandparents. So, which grandparent did that actual segment that descended to the grandchild descend from?
For people who are from the same core population on both parent’s side, close matches are often your only “sure thing” and beyond that, hopefully you have your parents (at least one parent) available to match against, because that’s the only way of even beginning to sort into family groups. This is known as phasing against your parents and while it’s a great tool for everyone to use – it’s essential to people who descend from endogamous groups. Endogamy makes genetic genealogy difficult.
In other cases, where you do have endogamy in your line, but only in one of your lines, endogamy can actually help you, because you will immediately know based on who those people match in addition to you (preferably on the same segment) which group they descend from. I can’t tell you how many rows I have on my spreadsheet that are labeled with the word “Acadian,” “Brethren” and “Mennonite.” I note the common ancestor we can find, but in reality, who knows which upstream ancestor in the endogamous population the DNA originated with.
Now, the bad news is that Ancestry runs a routine that removes DNA that they feel is too matchy in your results, and most of my Acadian matches disappeared when Ancestry implemented their form of population based phasing.
Identical by Population
There is sometimes a fine line between a match that’s from an ancestor one generation further back than you can go, and a match from generations ago via DNA found at a comparatively high percentage in a particular population. You can’t tell the difference. All you know is that you can’t assign that segment to an ancestor, and you may know it does phase against a parent, so it’s valid, meaning not IBC or identical by chance.
Yes, identical by population segment matching is a distinct problem with endogamy, but it can also be problematic with people from the same region of the world but not members of endogamous populations. Endogamy is a term for the timeframe we’re familiar with. We don’t know what happened before we know what happened.
From time to time, you’ll begin to see something “odd” happened where a group of segments that you already have triangulated to one ancestor will then begin to triangulate to a second ancestor. I’m not talking about the normal two groups for every address – one from your Mom’s side and one from your Dad’s. I’m talking, for example, when my Mom’s DNA in a particular area begins to triangulate to one ancestral group from Germany and one from France. These clearly aren’t the same ancestors, and we know that one particular “spot” or segment range that I received from her DNA can only come from one ancestor. But these segment matches look to be breaking that rule.
I created the example below to illustrate this phenomenon. Notice that the top and bottom 3 all match nicely to me and to each other and share a common ancestor, although not the same common ancestor for the two groups. However, the range significantly overlaps. And then there is the match to Mary Ann in the middle whose common ancestor to me is unknown.
Generally, we see these on smaller segment groups, and this is indicative that you may be seeing an identical by population group. Many people lump these IBP (identical by population) groups in with IBC, identical by chance, but they aren’t. The difference is that the DNA in an IBP group truly is coming from your ancestors – it’s just that two distinct groups of ancestors have the same DNA because at some point, they shared a common ancestor. This is the issue that “academic phasing” (as opposed to parental phasing) is trying to address. This is what Ancestry calls “pileup areas” and attempts to weed out of your results. It’s difficult to determine where the legitimate mathematical line is relative to genealogically useful matches versus ones that aren’t. And as far as I’m concerned, knowing that my match is “European” or “Native” or “African” even if I can’t go any further is still useful.
Think about this, if every European has between 1 and 4% Neanderthal DNA from just a few Neanderthal individuals that lived more than 20,000 years ago in Europe – why wouldn’t we occasionally trip over some common DNA from long ago that found its way into two different family lines.
When I find these multiple groupings, which is actually relatively rare, I note them and just keep on matching and triangulating, although I don’t use these segments to draw any conclusions until a much larger triangulated segment match with an identified ancestor comes into play. Confidence increases with larger segments.
This multiple grouping phenomenon is a hint of a story I don’t know – and may never know. Just because I don’t quite know how to interpret it today doesn’t mean it isn’t valid. In time, maybe its full story will be revealed.
ROH – Runs of Homozygosity
Autosomal DNA tests test someplace over 500,000 locations, depending on the vendor you select. At each of those locations, you find a value of either T, A, C or G, representing a specific nucleotide. Sometimes, you find runs of the same nucleotide, so you will find an entire group of all T, for example. If either of your parents have all Ts in the same location, then you will match anyone with any combination of T and anything else.
In the example above, you can see that you inherited T from both your Mom and Dad. Endogamy maybe?
Sally, although she will technically show as a match, doesn’t really “match” you. It’s just a fluke that her DNA matches your DNA by hopping back and forth between her Mom’s and Dad’s DNA. This is not a match my descent, but by chance, or IBC (identical by chance.) There is no way for you to know this, except by also comparing your results to Sally’s parents – another example of parental phasing. You won’t match Sally’s parents on this segment, so the segment is IBC.
Now let’s look at Joe. Joe matches you legitimately, but you can’t tell by just looking at this whether Joe matches you on your Mom’s or Dad’s side. Unfortunately, because no one’s DNA comes with a zipper or two sides of the street labeled Mom and Dad – the only way to determine how Joe matches you is to either phase against Joe’s parents or see who else Joe matches that you match, preferable on the same segment – in other words – create either a match or ICW group, or triangulation.
Segment Size
Everyone is in agreement about one thing. Large segments are never IBC, identical by chance. And I hate to use words like never, so today, interpret never to mean “not yet found.” I’ve seen that large segment number be defined both 13cM and 15cM and “almost never” over 10cM. There is currently discussion surrounding the X chromosome and false positives at about this threshold, but the jury is still out on this one.
Most medium segments hold true too. Medium segment matches to multiple people with the same ancestors almost always hold true. In fact, I don’t personally know of one that didn’t, but that isn’t to say it hasn’t happened.
By medium segments, most people say 7cM and above. Some say 5cM and above with multiple matching individuals.
As the segment size decreases, the confidence level decreases too, but can be increased by either multiple matches on that segment from a common proven ancestor or, of course, triangulation. Phasing against your parent also assures that the match is not IBD. As you can see, there are tools and techniques to increase your confidence when dealing with small segments, and to eliminate IBC segments.
The issue of small segments, how and when they can be utilized is still unresolved. Some people simply delete them. I feel that is throwing the baby away with the bathwater and small segments that triangulate from a common ancestor and that don’t find themselves in the middle of a pileup region that is identical by population or that is known to be overly matchy (near the center of chromosome 6, for example) can be utilized. In some cases, these segments are proven because that same small segment section is also proven against matches that are much larger in a few descendants.
Tim Janzen says that he is more inclined to look at the number of SNPs instead of the segment size, and his comfort number is 500 SNPs or above.
The flip side of this is, as David Pike mentioned, that the fewer locations you have in a row, the greater the chance that you can randomly match, or that you can have runs of heterozygosity.
No one in our discussion group felt that all small segments were useless, although the jury is still out in terms of consensus about what exactly defines a small segment and when they are legitimate and/or useful. Everyone of us wants to work towards answers, because for those of us who are dealing with colonial ancestors and have already picked the available low hanging fruit, those tantalizing small segments may be all that is left of the ancestor we so desperately need to identify.
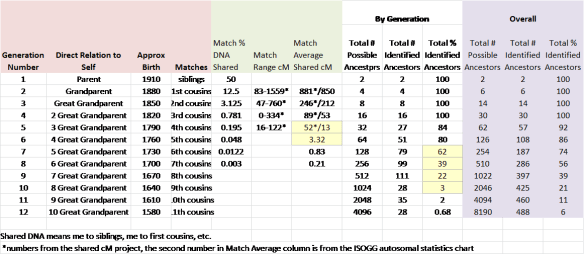
For example, I put together this chart detailing my matching DNA by generation. Interesting, I did a similar chart originally almost exactly three years ago and although it has seemed slow day by day, I made a lot of progress when a couple of brick walls fell, in particular, my Dutch wall thanks to Yvette Hoitink.
If you look at the green group of numbers, that is the amount of shared DNA to be expected at each level. The number of shared cMs drops dramatically between the 5th and 6th generation from 13 cM which would be considered a reasonable matching level (according to the above discussion) at the 5th generation, and 3.32 cM at the 6th generation level, which is a small segment by anyone’s definition.
The 6th generation was born roughly in 1760, and if you look to the white grouping to the right of the green group, you can see that my percentage of known ancestors is 84% in the 5th generation, 80% in the 6th generation, but drops quickly after that to 39, 22 and 3%, respectively. So, the exact place where I need the most help is also the exact place where the expected amount of DNA drops from 13 to 3.32 cM. This means, that if anyone ever wants to solve those genealogical puzzles in that timeframe utilizing genetic genealogy, we had better figure out how to utilize those small segments effectively – because it may well be all we have except for the occasional larger sticky segment that is passed intact from an ancestor many generations past.
From my perspective, it’s a crying shame that Ancestry gives us no segment data and it’s sad that 23andMe only gives us 5cM and above. It’s a blessing that we can select our own threshold at GedMatch. I’m extremely grateful that FTDNA shows us the small segment matches to 1cM and 500 SNPs if we also match on 20cM total and at least one segment over 7cM. That’s a good compromise, because small segments are more likely to be legitimate if we have a legitimate match on a larger segment and a known ancestor. We already discussed that the larger the matching segment, the more likely it is to be valid. I would like to see Family Tree DNA lower the matching threshold within projects. Surname projects imply that a group of people will be expected to match, so I’d really like to be able to see those lower threshold matches.
I’m hopeful that Family Tree DNA will continue to provide small segment information to us. People who don’t want to learn how to use or be bothered with small segments don’t have to. Delete is perfectly legitimate option, but without the data, those of us who are interested in researching how to best utilize these segments, can’t. And when we don’t have data to use, we all lose. So, thank you Family Tree DNA.
Coming Full Circle
This discussion brings us full circle once again to goals.
Goals change over time.
My initial reason for testing, the first day an autosomal test could be ordered, was to see if my half-brother was my half-brother. Obviously for that, I didn’t need matching to other people or triangulation. The answer was either yes or no, we do match at the half-sibling level, or we don’t.
He wasn’t. But by then, he was terminally ill, and I never told him. It certainly explained why I wasn’t a transplant match for him.
My next goal, almost immediately, was to determine which if either my brother or I were the child of my father. For that, we did need matching to other people, and preferably close cousins – the closer the better. Autosomal DNA testing was new at that time, and I had to recruit cousins. Bless those who took pity on me and tested, because I was truly desperate to know.
Suffice it to say that the wait was a roller coaster ride of emotion.
If I was not my father’s child, I had just done 30+ years of someone else’s genealogy – not a revelation I relished, at all.
I was my father’s child. My brother wasn’t. I was glad I never told him the first part, because I didn’t have to tell him this part either.
My goal at that point changed to more of a general interest nature as more cousins tested and we matched, verifying different lineages that has been unable to be verified by Y or mtDNA testing.
Then one day, something magical happened.
One of my Y lines, Marcus Younger, whose Y line is a result of a NPE, nonparental event, or said differently, an undocumented adoption, received amazing information. The paternal Younger family line we believed Marcus descended from, he didn’t. However, autosomal DNA confirmed that even though he is not the paternal child of that line, he is still autosomally related to that line, sharing a common ancestor – suggesting that he may have been born of a Younger female and given that surname, while carrying the Y DNA of his biological father, who remains unidentified.
Amazingly, the next day, a match popped up that matched me and another Younger relative. This match descended not from the Younger line, but from Marcus Younger’s wife’s alleged surname family. I suddenly realized that not only was autosomal DNA interesting for confirming your tree – it could also be used to break down long-standing brick walls. That’s where I’ve been focused ever since.
That’s a very different goal from where I began, and my current goal utilizes the tools in a very different way than my earlier goals. Confidence levels matter now, a great deal, where that first day, all I wanted was a yes or no.
Today, my goal, other than breaking down brick walls, is for genetic genealogy to become automated and much easier but without taking away our options or keeping us so “safe” that we have no tools (Ancestry).
The process that will allow us to refine genetic genealogy and group individuals and matches utilizing trees on our desktops will ultimately be the key to unraveling those distant connections. The data is there, we just have to learn how to use it most effectively, and the key, other than software, is collaboration with many cousins.
Aside from science and technology, the other wonderful aspect of autosomal DNA testing is that is has the potential to unite and often, reunite families who didn’t even know they were families. I’ve seen this over and over now and I still marvel at this miracle given to us by our ancestors – their DNA.
So, regardless of where you fall on the goals and matching confidence spectrum in terms of genetic genealogy, keep encouraging others to test and keep reaching out and sharing – because it takes a village to recreate an ancestor! No one can do it alone, and the more people who test and share, the better all of our chances become to achieve whatever genetic genealogy goals we have.
______________________________________________________________
Disclosure
I receive a small contribution when you click on some of the links to vendors in my articles. This does NOT increase the price you pay but helps me to keep the lights on and this informational blog free for everyone. Please click on the links in the articles or to the vendors below if you are purchasing products or DNA testing.
Thank you so much.
DNA Purchases and Free Transfers
- Family Tree DNA
- MyHeritage DNA only
- MyHeritage DNA plus Health
- MyHeritage FREE DNA file upload
- AncestryDNA
- 23andMe Ancestry
- 23andMe Ancestry Plus Health
- LivingDNA
Genealogy Services
Genealogy Research
- Legacy Tree Genealogists for genealogy research
























This is the best article on DNA genealogy I have ever read! It pulls together all the bits and pieces I’ve been wallowing around in. I’m recommending it to anyone I discuss DNAG with!
Thank you. You’re not wallowing alone:)
Roberta, while I would agree with Blaine’s chart about the order of confidence, I don’t agree with the chart you have under it. How often have you seen pseudo-triangulation fail?
I would say that, if you’re working with segments over 10 cM and you have more than a couple of people in a pseudo-triangulated group who all ICW one another the odds that the pseudo-triangulation is mirroring a true triangulation are in the high-90 percentile – at least in my experience. This is particularly true if you think about the fact that, for it to fail, the segment you see in a supposed TG has to be on your other parental chromosome of the pair. For that to happen that match would have to just happen to coincidentally match every one of the other members of the pseudo-TG in some other way. Unless you are Ashkenazim, that’s not very likely to happen and it’s pretty easy to visualize in ADSA because of the scarcity of ICW bricks in the “field” away from the central diagonal line.
So assigning “Medium” to the confidence for something that most of the time is close to 100% confidence seems a little misleading. Especially when you put it at the same level of confidence as an Ancestry Shakey Leaf match (where I have found examples that failed).
I’m not quibbling about where to slot these things. It’s more than I don’t think you can quantify confidence like this – at least in absolute terms. It really depends on the circumstances at hand which can vary. A close Ancestry match is going to have a much higher confidence level than a more distant one and a close Ancestry match is more reliable than a distant FTDNA match.
Don
I did have trouble exactly figuring out where to slot things. The group you’re referring to are in order, I think, not equal. And match groups differ at different levels of cMs, which is why I didn’t rank them higher. It varies and it has to do with the circumstances. I think the real message here is that you have to evaluate everything about the circumstances of the match before assigning it your own level of confidence.
Yep! I’ve found that there are no absolutes in DNA (not even with true triangulation). There is always a non-zero probability on both sides of the equation, although it can be pretty skewed in one direction or the other. It’s a lot like traditional genealogy in that a single source isn’t absolute proof – how many errors have you and I seen in censuses and death certificates, for example? It pays to have multiple corroborating pieces of evidence – DNA and traditional. That’s why I like to have multiple DNA matches that all tell me the same thing before I start to really believe it!
I agree 100%! Absolute rules would be a lot easier:)
Don
It depends …if you have a mildly endogamous group pseudo-triangulation can and will fail (my Norwegians) but if for normal multi admixed Americans it is fine. The DNAadoption folk give it 90-99%.
Kitty
Great educational piece. I thank you for taking the time to pull this all together. I am a bit puzzled why you make no mention of the fantastic Autosomal DNA tool Genome Mate Pro. IMHO, it allows one to pull together and analyze in one product the data which you cover in different spreadsheets. However, I certainly understand after all of these years the concept of “different strokes for different folks”. I have shared your article widely, and it now resides in my Evernote reference file. I look forward to your next analysis!
Those of us, like Roberta, who have been doing this a long time, have built our spreadsheets and hesitate to convert to an all-in-one product. I have heard nothing but good things about genomemate, but am used to my own ways. Someday I may find time to try it … (did I answer for you Roberta?)
Great article, Roberta! Thanks.
I enjoyed the article, Roberta. I’m starting to wonder – are there places in the world that weren’t endogamous 200+ years ago?
Good question. Probably not many.
Many thanks for this one!! I’m a “small seg” believer… pat davis in OH
Wonderful article, thank you Roberta. Now I understand your interest in small segments.
One small point, those of us who have siblings and parents tested can use each other to triangulate matches at family tree DNA. The best example is when A and B match Dad on the same segment and my brother and Dad match person A but brother does not match B. Meanwhile I match B. Likely A and B are NOT a match on that segment. Conversely when I match A and B as does Dad on a specific segment then we have triangulation, I got that segment from Dad. My blog post on triangulation dicusses this technique. These of course are all medium to large segments that I work with 🙂
Kitty
Will you please post the link to your article, Kitty. Thanks much.
Sure Roberta, here is my triangulation article
http://blog.kittycooper.com/2015/02/triangulation-proving-a-common-ancestor/
using close family to triangulate at ftDNA is discussed in the last few paragraphs
I’ve found this strategy to be exceedingly useful.
Thanks so much for laying everything out so clearly, though I agree with Don Worth re pseudo-triangulation. I love the way you’ve presented and explained the various options for working with atDNA – and without belittling or excluding any of the competing companies. 🙂
Excellent article, Roberta.
The issue of what constitutes a match at the different companies, is a source of confusion. You said in this article that at FTDNA there has to be at least one segment over 7 cM, but the ISOGG chart says that it has to be at least 7.69 cM. I tried to find this information on the FTDNA site, and the only reference I could find was that it had to be at least 5.5 cM. My experience with a person who matches my two sisters is that I think that the 7.69 cM value is correct. This is because this person matches both sisters on AncestryDNA but only the one with a 24.6 cM segment on FTDNA. From GEDmatch I found that the other sister’s largest segment is only 7.4 cM, and presumably that is why she does not show as match on FTDNA. I did a matrix comparison on FTDNA and again only one sister shows as a match. It is a pity that the minimum segment size is not a parameter that one can change when needed, as far as the initial match is concerned.
You can easily tell by downloading all of your matches from FTDNA and then looking at matches that have 7.0 or between 7.0 and 7.69. Then look at the other cMs for those matches. I did this and I have several matches where they have segments between 7.0 and 7.69 and that is the largest single segment, although there are several smaller segments. Check your own data to see what you find.
I tried sorting my FTDNA matches according to longest block and then going to my last page of matches. They all had 7.69 cM or greater. Not sure how you could find matches with smaller largest segment.
I downloaded everything into a spreadsheet.
I did try downloading and got the same result – smallest block was 7.69 cM
I wonder if that has changed then. My download results are a composite over time. Interesting. I wonder why 7.69. Seems like a very odd number.
I wondered also about it having changed over time. My results are from 2013, but I imagine that yours are much older than that.
In any case let me say once again that this is a very educational article.
Thanks.
Another magnificent piece, Roberta! You selflessly take time and trouble to educate those of us eager to learn, albeit little by slowly. The more I understand, the more I CAN understand. Over and over, you guide me into my next level of understanding. Six months ago, the title of this blog would have baffled me! You are a fine teacher, steering us into our own ability to help others, which begins by explaining to our matches why they should upload to Gedmatch. Those folks, in turn, see the value and hopefully encourage their matches to do the same. We ARE very much in this together.
See, you think it’s selfless, but it’s not:) Thank you for your kind comments. You’re right, we are all in this together. We win together, or not.
I use GenomeMate to track people who share segments 10cM and above with me, downloading from FTDNA and Gedmatch. I also use DNAGedcom to look at all the people with tiny segments, that I have downloaded from FTDNA. I haven’t used their beta tool for Gedmatch yet. This way I have different datasets to work with. I do not have the skills to set up Excel spreadsheets.
This blog has helped me understand genetic genealogy much better. Even though I have been doing traditional genealogy for 39 years, it is a steep learning curve for genetic genealogy.
Another great article Roberta
I have a question.
My wife in Fdtna ahs a 5thcousin -remote cousin with a shared CM of 43.73.
By birth certificates I have found this union and shared information with this person, but this person ended up being 9th cousin……..the question is………is 43.73 CM high to be only a 9th cousin?
regards
Yes, it is high but that is why averages and guidelines are only that.
Re the small segments, Paul Rakow just put together some useful data analysis that includes pointing out that “Only 32% of 5th cousins have any DNA shared with you, only 11% have
a match that’s longer than 10 cM.” In other words, it’s not that all those distant cousins can be assumed to share segments of 3.32 cM (or whatever as the distance increases), it’s that that that would be the average and instead of looking at the average, we need to drop all the non-sharers from the calculation and instead look at the average of the actual shared cM, which will be a more useful guide.
In other words, if 32% of 5th cousins share DNA with you, you will be noticing the 11% with a match longer than 10cM and also those who have a match between 7-10cM (which, added in, probably gets us close to that whole 32%).
I want to see all the tiny segments for my matches, but that is because I’ve found that read errors and whatnot often lop a piece off a segment from a known relative, and because sure, it’s possible that several known cousins may share some small segments that can be proven to be IBD. I am not hunting for people who don’t have a segment of at least 7cM in common with a known family member.
Another remarkably informative article, Roberta! You set a very high standard in helpfulness and productivity.
You mentioned that “The classic definition of triangulation is when three people, A, B and C, all match each other on the same segment and share a known, identifiable common ancestor.” If A is oneself, how distantly related to each other should B and C be for a high-confidence triangulation? For example, if B and C are siblings, I would think this would not be considered sufficient triangulation.
Thanks again,
Jim
Hi Jim, Siblings and parent can’t be used for triangulation. All that does is to verify that their common parent carried that DNA, so that’s really not triangulation. I’m more comfortable with at least first cousins, but with more people added to the group would be even more confident yet.
Wonderful article Roberta. I am so happy to know I am not the only one with a question about chromosome 6 just before the centromere. It is also helpful to know why some KNOWN relatives do not show up in my Ancestry DNA matches – but in my FTDNA matches; I am sure they are to matchey. Thank you so much you have saved me worry and work.
Thanks, Roberta, for another highly educational article. I had previously figured out that triangulation was the “gold standard” but still had lots of uncertainty about how to evaluate the other sorts of matches. I also have to cope with colonial endogamy on the paternal side of my tree which has added to my confusion so I found those paragraphs on endogamy and IBP especially valuable.
Roberta, See at one of your X chromo blogs that you have… 28 – Agnes Craven is the last slot on the chart, but not the last in the line. Her father was Col. Robert Craven born 1696 in Delaware and was well to do. His mother is unknown. Robert’s wife was Mary Harrison, born in Oyster Bay, New York to Isaiah Harrison and Elizabeth Wright… I have DNA matches w/ the Harrison fam… I’m working on a DAVIS brick wall, possibly from Oyster Bay, L.I. NY…
You mentioned GEDmatch… would you pls email your kit #s. Thank you, Pat Davis in OH kit#F244618
My kit number is F6656.
Roberta,
I am just repeating the obvious, that this is the most educating article to date. You are my favorite blogger of all–that includes photography, knitting, painting, etc.
Without your articles I would not have stuck to learning all about DNA and making one million spreadsheets to accommodate all the information.
Thank you again for all your education.
klmmer
Thank you!
Although I agree that Roberta’s articles are real “pearls” of DNA knowledge, there is one issue I am struggling to understand: the “triangulation”. If A matches me at a specific segment of the chromosome 11 for instance, and B matches me at THE SAME SEGMENT, how is it possible that A does not match B? After all there is a fundamental dogma in Math, that if A = C, and B = C, then A = B!
henryg
It’s not a match equation. The reason is because you, and they, all have two segments. Remember – you have one from Mom and one from Dad. They can match you on the same “segment” number, but they can actually be matching you on different sides of your DNA. Think of it as two streets with the same house numbers. From just the house numbers, you can’t tell which street.
Dear Roberta,
What you say is very true, however, if FTDNA does not do “phasing” there is no way to tell which branch of the two are matching, and A and B would still “match”, even, if on different branches (which they would not know). Any way to solve this problem?
Regards,
Henryk Gruder
If A and B don’t match each other, then you know they are from opposite sides. If you have a parent test, then it’s easy to tell. Academic phasing would not help this situation – only parental phasing which you can do by seeing who matches your parents, if they have tested. Otherwise, you have to wait until you get a recognizable match on this segment (and to one of the other people) to someone with whom you can identify your common ancestor.
Yes. It makes sense. Thanks.
HG
Dear Roberta,
Can one use his child as a second part in triangulation (himself would be one)? Or it is just like using his own data twice?
Many thanks,
Henryk Gruder
No, two closely related people can’t both be used in triangulation. Your child only has the DNA you (and their other parent) gave them, so they are not a proof point, but a subset of you genetically.
Dear Roberta,
Now a problem related to X chromosome. I read your blogs related to it with a keen interest, but, being just a mathematician, I still struggle with some concepts.
There is a female person, with the same surname as mine (I am male ), who, according to Gedmatch shares 7.3 cM on chromosome 1, but 196 common block on 23 / X chromosome. She uses her grandfather surname. In spite of that FTDNA does not show her as my “match” (perhaps of endogamy problem, we are Ashkenazi Jews). In one of your blogs I have found that 195 cM could be shared between a grandma and granddaughter. 196 is significant, on the other hand not a “match”. How should I understand that?
Thanks for your help,
Henryk Gruder
FTDNA does not show you as a match unless you match on 20cM of other DNA total.
So a block of 196 cM in chromosome 23 does not mean anything??
That’s not what I said. I said FTDNA doesn’t show an X match unless you have more than 20cM of other matches total. You asked why your match wasn’t showing.
It means you have a match and you need to use the specific X related pedigree chart to attempt to find a common ancestor. The X chromosome can and is passed intact sometimes for generations, so you may or may not be able to find that common ancestor.
Another question (sorry, being a “newbie’ in genetic genealogy I have plenty):
In your (excellent) article “…DNA matching confidence..”, page 26 there is a table containing some examples of data, among them sizes of shared cM. Do you mean total shared, or max. block (FTDNA shows both)?
Thanks,
Henryk Gruder
I don’t use max block for anything.
(About X chromosome again).
Yes, I have read about “the uruly X chromosome”, and understand that having shared 196 cM (100%, if i correctly understood your article?) with my female (I am male) relative (?) does not tell the whole story, however, perhaps, some conclusions can be drawn (even in the endogamy – Ashkenazi situation) ?
I have five DNA matches although one is very small and I know we all share a common ancestor about 5-7 generations back. However, I am not able to find a matching DNA segment between all of us. What does this mean? Thanks!
It means you don’t share any common segments from that ancestor. Not terribly unusual for matches from quite a distance back in time.
Pingback: 11th International Conference on Genetic Genealogy – Sunday | Ancestor Central
I appreciate your addressing the smaller segments, Roberta. I have several brick walls in the 6-7 generation bracket that i am dealing with, and these borderline segments are very important to my research in these areas and I have had success using them.
Getting matches to upload to gedmatch is paramount but many of these matches drop off of the 1500 match limit of the gedmatch one/many tool. I recently copied my one/many results to a spreadsheet and then sorted the original results on gedmatch “up” then “down” by account numbers and added each of these results to my spreadsheet. After deleting duplicate account numbers, I found an additional 2500 matches that fell within the gedmatch 7/500 default setting for the one/many tool. I still missed many more matches as the sorting failed to capture many more ancestryDNA and FTDNA matches..
As the number of viable matches increase for each of us Gedmatch should increase their results to include all matches that meet their criteria for the one/many tool or they should allow results to be obtained by testing site.
You can get all of your matches if you use the Matching Segments Search tool (Tier 1). My wife has 2700 matches and all of them (and their segments) show up in the MSS. Of course, that requires a subscription to GEDMATCH.
Pingback: The Best and Worst of 2015 – Genetic Genealogy Year in Review | DNAeXplained – Genetic Genealogy
Pingback: We Match…But Are We Related? | DNAeXplained – Genetic Genealogy
Pingback: The Ancestry 200 | DNAeXplained – Genetic Genealogy
Pingback: Concepts – How Your Autosomal DNA Identifies Your Ancestors | DNAeXplained – Genetic Genealogy
Pingback: Curtis Benjamin Lore (1856-1909), Devilishly Handsome Rogue, 52 Ancestors #113 | DNAeXplained – Genetic Genealogy
Pingback: Concepts – Identical by…Descent, State, Population and Chance | DNAeXplained – Genetic Genealogy
Pingback: Concepts – CentiMorgans, SNPs and Pickin’ Crab | DNAeXplained – Genetic Genealogy
Pingback: Concepts – Parental Phasing | DNAeXplained – Genetic Genealogy
Thanks for your informative, interesting and entertaining blogs.
I have a question for which I cannot find the answer after searching your blogs and other on line sources: If small segments in a GEDMatch triangulation have exactly the same start and/or end point, (as contrasted to just overlapping), does this tend to increase the reliability of a IBD match for that particular segment?
I’m not positive of the answer. Each vendor handles boundaries differently. So an exact boundary may be a reflection of any address within a specific range. I don’t know how GedMatch handles this. In smaller segments, I would be more suspicious of non-exact boundary matches.
Thanks for the fast reply. My (hopeful) thinking, in being forced to work with some small segment triangulation, is that the same start and/or end point (if they are, indeed, exact), in the random course of the chromosome recombining events is less likely to be by chance than by common source of the segment. I shall follow up with GEDMatch. Gary Morrison
Gary, from what I have read most of the companies process matches in “bundles” of small segments of DNA (100 SNPs or so) (Ancestry calls them “windows” in some of their white papers). In any case, this leads to exactly the same starting base pair location and/or ending location in some cases. I’ve been told this is not significant of anything other than as an artifact of the matching process. The key is in how much overlap (measured in centimorgans) there is between matching segments in a triangulated group.
Thanks, Don. I just found a FAQ in FTDNA’s website that answers the question exactly as you have so that makes it official … no significance to an exact match on start or end point for overlapping segments. Darn. Cheers, Gary
Remember that each vendor handles things a bit differently.
Roberta, right you are. I had not considered in my response to Don that it is available for all to see. In my case, the testing was done by FTDNA, so I took their information as “the” answer but for others who test with a different vendor the answer might well be different.
Thanks for providing this forum to help us all along on this fascinating but sometimes mystifying journey. – Gary
Your article above is THE BEST i have seen explaining DNA genealogy for people. You really get it and you write well.
Yes, Ancestry’s NAD feature is falsely named. Most NADs are so easily dismissed that NAD is the sort of thing that leads people to refer to artificial intelligence as artificial stupidity. (I realize that AI is not used here). Only the name is at fault, though. These can be VERY useful because they identify groups of cousins we might not have noticed as being related to each other. In one case where I matched just two out of a dozen in the circle, two who were more closely related to each other than to the rest in the circle, and i noticed that the two had a common ancestor (not the NAD) with a surname spelled a little differently from a surname in my tree but probably connecting to my line in NC before 1800. The NAD was wrong, but the clue was really helpful.
What Ancestry needs is a cousins group finder, CGF, that identifies groups and tells us how some people in the group are related to each other if their public trees connect. Why must we ploddingly discover what Ancestry’s computers already know? You are so right that software SHOULD be able to do this and much more… Someday.
Pingback: Which DNA Test is Best? | DNAeXplained – Genetic Genealogy
Pingback: Concepts – Why Genetic Genealogy and Triangulation? | DNAeXplained – Genetic Genealogy