What, exactly, is an autosomal DNA match?
Answer: It’s Relative
I’m sorry, I just had to say that.
But truthfully, it is.
I know this sounds like a very basic question, and it is, but the answer sometimes isn’t as straightforward as we would like for it to be.
Plus, there are differences in quality of matches and types of matches. If you want to sigh right about now, it’s OK.
We’ve talked a lot about matching in various recent articles. I have several people who follow this blog religiously, and who would rather read this than, say, do dishes (who wouldn’t). One of our regulars recently asked me the question, “what, exactly, is a match and how do I tell?”
Darned good question and I wish someone had explained this to me so I wouldn’t have had to figure it out.
In the computer industry, where I spent many years, we have what we call flow charts or wernier diagrams which in essence are logic paths that lead to specific results or outcomes depending on the answers at different junctions.
I had a really hard time deciding whether to use the beer decision-making flow chart or the procrastinator flow chart, but the procrastinator flow chart was just one big endless loop, so I decided on the beer.
What I’m going to do is to step you through the logic path of finding and evaluating a match, determining whether it’s valid, identical by descent or chance, when possible, and how to work with your matches and what they mean.
Let me also say that while I use and prefer Family Tree DNA, these matching techniques are universal and apply to results from 23andMe as well, but not for Ancestry who gives you no browser or tools to compare your DNA to anyone else. So, you can’t compare your results at Ancestry.
Comparing DNA results is the lynchpin of genetic genealogy. You’re dead in the water without it. If you have tested at Ancestry, you can always transfer your results to Family Tree DNA, where you do have tools, and to GedMatch as well. You’re always better, in terms of genealogy, to fish in as many ponds as possible.
Before we talk about how to work with matches, for those who need to figure out how to find matches at Family Tree DNA and 23andMe, I wrote about that in the Chromosome Browser War article. This article focuses on working with matching DNA after you have found that you are a match to someone – and what those matches might mean.
Matching Thresholds
All autosomal DNA vendors have matching thresholds. People who meet or exceed those thresholds will be shown on your match list. People who do not meet the initial threshold will not be considered as a match to you, and therefore will not be on your match list.
Currently, at Family Tree DNA, their match threshold to be shown as a match is about 20cM of total matching DNA and a single segment of about 7.7cM with 500 SNPs or over. The words “about” are in there because there is some fuzziness in the rules based on certain situations.
After you meet that criteria and you are shown as a match to an individual, when you download your matching data, your matches to them on each chromosome will be shown to the 1cM and 500 SNP level
At 23andMe, the threshold is 7cMs/700 SNPs for the first segment. However, 23andMe has an upper limit of people who can match you at about 1000 matches. This can be increased by the number of people you are communicating or sharing with. However, your smallest matches will be dropped from your list when you hit your threshold. This means that it’s very likely that at least some of your matches are not showing if you have in excess of 1000 matches total. This means that your personal effective cM/SNP match threshold at 23andMe may be much higher.
Step 1 – Downloading Your Matching Segments
For this comparison, I’m starting with two fresh files from Family Tree DNA, one file of my own matches and one of my mother’s matches. My mother died before autosomal DNA testing was available, so her results are only at Family Tree DNA (and now downloaded to GedMatch,) because her DNA was archived there. Thank you Family Tree DNA, 100,000 times thank you!!!
At Family Tree DNA, the option to download all matches with segment information is on the chromosome browser tab, at the top, at the right, shown below.
If you have your parents DNA available to test and it hasn’t been tested, order a kit for them today. If either or both parents have been tested, download their results into the same spreadsheet with yours and color code them in a way you will understand.
In my case, I only have my mother’s results, and I color coded my matches pink, because I’m the daughter. However, if I had both parents, I might have colored coded Mother pink and Dad blue.
Whatever color coding you do, it’s forever in your master spreadsheet, so make a note of what it is. In my case, it’s part of the match column header. Why is it in my column header? Because I screwed up once and reversed them in a download.
Step 2 – Preparing and Sorting Your Spreadsheet
In my master DNA spreadsheet, I have the following columns,
The green cell matches are matches to me from 23andMe. My cousin, Cheryl also tested at 23andMe before autosomal testing was offered at Family Tree DNA.
The Source column, in my spreadsheet, means any source other than FTDNA. The Ignore column is an extraneous number generated at one time by downloads. I could delete that column now.
The “Side” column is which side the match is from, Mom or Dad. Mom’s I can identify easily, because I have her DNA to compare to. I don’t identify a match as Dad’s without having identified an ancestral line, because I don’t have his DNA to compare to.
And no, you can’t just assume that if it doesn’t match Mom, it’s an automatic match to Dad because you may have some IBS, identical by chance, matches.
The Common Ancestors/Comments column is just that. I include things like when I e-mailed someone, if the match is triangulated and if so, with whom, etc.
In my master spreadsheet, the first “name” column (of who tested) is deleted, but I’ve left it in the working spreadsheet (below) with my mother for illustration purposes. That way, neither of us has to remember who is pink!
Step 3 – Reviewing IBD and IBS Guidelines
If you need a refresher on, phasing, IBD, identical by descent, IBS which can mean either identical by chance or identical by population, it would be a good time to read or reread the article titled How Phasing Works and Determining IBD Versus IBS Matches.
Let’s briefly review the IBD vs IBS guidelines, because we’ll be applying them in this article.
Identical by Chance – Can be determined if an individual you match does not match to one of your parents, if parents are available. If parents are not available for matching, IBS by chance segments won’t triangulate with other known genealogical matches on a common segment.
Identical by Descent – Can be suggested if a common ancestor (or ancestral line) can be determined between any two people who are not known relatives. If the two people are known close relatives, and their DNA matches, identical by descent is proven. IBD can be proven with previously unknown family or genealogical matches when any three people descending from that same ancestor or ancestral line all match each other on the same segment of DNA. Three way matching is called triangulation.
Identical by Population – Can be determined when multiple people triangulate with you on a specific segment of DNA, but the triangulated groups are from proven different lineages and are not otherwise related. This is generally found in smaller segments from similar regions of the world. Identical by population is identical by descent, but the ancestors are so far back in time that they cannot be determined and may contribute the same DNA to multiple lineages. This is particularly evident in Jewish genealogy and other endogamous groups.
Step 4 – Determining Parental Side and IBS by Chance
The first thing to do, if you have either or both parents, is to determine whether your matches phase to your parents or are IBS by chance.
In this context, phasing means determining whether a particular match is to your father’s side of the family or to your mother’s side of the family.
Remember, at every address in your DNA, you will have two valid matches to different lines, one from your mother and one from your father. The address on your DNA consists of the chromosome number which equates to the street name, and then the start and end locations, which consists of a range of addresses on that street. Think of it as the length of your property on the street.
First, let’s look at my situation with only my mother’s DNA for comparison.
It’s easy to tell one of three things.
- Do mother and I both match the person? If so, that means that DNA match is from mother’s side of the family. Mark it as such. They are green, below.
- If the individual does not match me and mother, both, and only matches me, then the match is either on my father’s side or it’s IBS by chance. Those matches are blue below. Because I don’t have my father’s DNA, I can’t tell any more at this step.
- Notice the matches that are Mom’s but not to me. That means that I did not receive that DNA from Mom, or I received a small part, but it’s not over the lowest matching threshold at Family Tree DNA of 1cM and 500 SNPs.
In this next scenario, you can see that mother and I both match the same individual, but not on all segments. I selected this particular match between me, my mother and Alfred because it has some “problems” to work through.
The segments shown in green above are segments that Mom carries that I don’t. This means that I didn’t receive them from mother. This also means they could be matching to Alfred legitimately, or are IBS by chance. I can’t tell anything more about them at this point, so I’ve just noted what they are. I usually mark these as “mother only” in my master spreadsheet.
The first of the two green rows above show a match but it’s a little unusual. My segment is larger than my mothers. This means that one of five things has happened.
- Part of this segment is a valid match. At the end, where we don’t match, the match extends IBS by chance a bit at the end, in my case, when matching Alfred. The valid match portion would end where my mother’s segment ends, at 16,100,293
- There is a read error in one of the files.
- The boundary locations are fuzzy, meaning vendor calculations like ‘healing’ for no calls, etc..
- I also match to my father’s line.
- Recombination has occurred, especially possible in an endogamous population, reconnecting identical by population segments between me and Alfred at the end of the segment where I don’t match my mother’s segment, so from 16,100,293 to 16,250,884.
Given that this is a small segment, the most likely scenario would be the first, that this is partly valid and partly IBS by chance. I just make the note by that row.
The second green segment above isn’t an exact match, but if my segment “fits within” the boundaries of my mother’s segments, then we know I inherited the entire segment from her. Once again, my boundaries are off a bit from hers, but this time it’s the beginning. The same criteria applies as in 1-5, above.
The green segments above are where I match Alfred, but my mother does not. This means that these segments are either IBS by chance or that they will match my father. I don’t know which, so I simply label them. Given that they are all small segments, they are likely IBS by chance, but we don’t know that. If we had my father’s DNA, we would be able to phase against him, too, but we don’t.
Now, if I was to leave this discussion here, you might have the impression that all small segment matches have problems, but they don’t. In fact, here’s a much more normal “rea life” situation where mother and I are both matching to our cousin, Cheryl, Mom’s first cousin. These matches include both large and small segments. Let’s take a look and see what we can tell about our matches.
Roberta and Barbara have a total of 83 DNA matches to Cheryl.
Some matches will be where Barbara matches Cheryl and Roberta doesn’t. That’s normal, Barbara is Roberta’s mother and Roberta only inherits half of Barbara’s DNA. These rows where only Barbara, the mother, matches Cheryl are not colorized in the Start, End, cM and SNP columns, so they show as white.
Some matches will be exact matches. That too is normal. In some cases, Barbara passes all of a particular segment of DNA to Roberta. These matches are colored purple.
Some of these matches are partial matches where Roberta inherited part of the segment of DNA from Barbara. These are colored green. There are two additional columns at right where the percentage of DNA that Roberta inherited from Barbara on these segments is calculated, both for cM and SNPs.
Some of the matches are where Roberta matches Cheryl and Barbara doesn’t. Cheryl is not known to be related to Roberta on her father’s side, so assuming that statement is correct, these matches would be IBS, identical by state, meaning identical by chance and can be disregarded at legitimate matches. These are colored rust. Note that most of these are small segments, but one segment is 8.8cM and 2197 SNPs. In this case, if this segment becomes important for any reason, I would be inclined to look at the raw data file of Barbara to see if there were no calls or a problem with reads in this region that would prevent an otherwise legitimate match.
Let’s look at how these matches stack up.
| Number | Percent (rounded) | Comment | |
| Exact Matches | 26 | 31 | 100% of the DNA |
| Barbara Only | 20 | 24 | 0% of the DNA |
| Partial Matches | 29 | 35 | 11-98% of the actual DNA matches |
| Roberta Only (IBS by chance) | 7 | 8 | Not a valid match |
I think it’s interesting to note that while, on the average, 50% of the DNA of any segment is passed to the child, in actuality, in this example of partial inheritance, meaning the green rows, inheritance was never actually 50%. In fact, the SNP and cM percentages inherited for the same segment varied, and the actual amounts ranged from 11-98% of the DNA of the parent being inherited by the child. The average of these events was 54.57143 (cM) and 54.21429 (SNPs) however.
On top of that, in 13 (26 rows) instances, Roberta inherited all of Barbara’s DNA in that sequence, and in 20 cases, Roberta inherited none of Barbara’s DNA in that sequence.
This illustrates that while the average of something may be 50%, none of the actual individual values may be 50% and the values themselves may include the entire range of possibilities. In this case, 11-98% were the actual percentage ranges for partial matches.
Matching Both Parents
I don’t have my father’s DNA, but I’m creating this next example as if I did.
Matches to mother are marked in green.
I have two matches where I match my father, so we can attribute those to his side, which I’ve done and marked in orange.
The third group of matches to me, at the bottom, to Julio, Anna, Cindy and George don’t match either parent, so they must be IBS by chance.
I label IBS by chance segments, but I don’t delete them because if I download again, I’ll have to go through this same analysis process if I don’t leave them in my spreadsheet
Step 5 – How Much of the DNA is a Match?
One person asked, “exactly how do I tell how much DNA is matching, especially between three people.” That’s a very valid question, especially since triangulation requires matching of three people, on the same segment, proven to a common ancestral line.
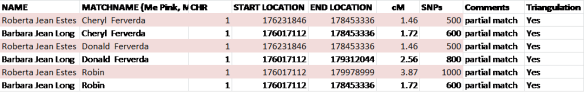
Let’s look at the match of both me and my mother to Don, Cheryl and Robin.
In this example, we know that Don, Cheryl and Robin all match me on my mother’s side, because they all three match me and my mother, both on the same segment.
How do we determine that we match on the same segment?
I have sorted this spreadsheet in order of end location, then start location, then chromosome number so that the entire spreadsheet is in chromosome order, then start location, then end location.
We can see that both mother and I match Cheryl partially on this segment of chromosome 1, but not exactly. The start location is slightly different, but the end location matches exactly.
The area where we all three match, meaning me, Mom and Cheryl, begins at 176,231,846 and ends at the common endpoint of 178,453,336
On the chart below, you can see that mother and I also both match Don, Cheryl’s brother, on part of this same segment, but not all of the same segment.
The common matching areas between me, Mom and Don begins at 176,231,846 and ends at 178,453,336.
Next, let’s look at the third person, Robin.
Mom and I both match Robin on part of this same overlapping segment as well. Note that my segment extends beyond Mom’s, but that does not invalidate the portion that does match between Robin, Mom and I.
Our common match area begins at the same location, but ends at 178,453,336, the same location as the common end area with Don and Cheryl
Step 6 – What Do Matches Mean? IBD vs IBS in Action
So, let’s look at various types of matches and what they tell us.
Looking at our matching situation above, let’s apply the various IBD/IBS rules and guidelines and see what we have
1. Are these matches identical by chance? No. How do we know?
a. Because they all match both me and a parent.
2. Are these matches identical by descent? Yes. How do we know?
a. Because we all match each other on this segment, and we know the common ancestor of Cheryl, Don, Barbara and me is Hiram Ferverda and Evaline Miller. We know that Robin descends from the same ancestral Miller line.
3. Are these matches identical by population. We don’t know, but there is no reason at this point to think so. Why?
a. Because looking at my master spreadsheet, I see no evidence that these segments are also assigned to other lineages. These individuals are also triangulated on a large number of other, much larger, segments as well.
4. Are these matches triangulated, meaning they are proven to a common ancestor? Yes. How do we know?
a. Documented genealogy of Hiram Ferverda and Evaline Miller. Don, Barbara, Cheryl and me are known family since birth.
b. Documented genealogy of Robin to the same ancestral family, even though Robin was previously unknown before DNA matching.
c. Even without the documented genealogy, Robin matches a set of two triangulation groups of people documented to the same ancestral line, which means she has to descend from that same line as well.
In our case, clearly these individuals share a common ancestor and a common ancestral line. Even though these are small segments on chromosome 1, there are much larger matching segments on other chromosomes, and the same rules still apply. The difference might be at some point smaller segments are more likely to be identical by population than larger segments. Larger segments, when available, are always safer to use to draw conclusions. Larger groups of matching individuals with known common genealogy on the same segments are also the safest way to draw conclusions.
Step 7 – Matching With No Parents
Sometimes you’re just not that lucky. Let’s say both of your parents have passed and you have no DNA from them.
That immediately eliminates phasing and the identical by chance test by comparing to your parents, so you’ll have to work with your matches, including your identical by chance segments.
A second way to “phase” part of your DNA to a side of your family is by matching with known cousins or any known family member.
In the situation above, matching to Cheryl, Don and Robin, let’s remove my mother and see what we have.
In this case, I still match to both of my first cousins, once removed, Cheryl and Don. Given that Cheryl and Don are both known cousins, since forever, I don’t feel the need for triangulation proof in this case – although the three of us are triangulated to our common ancestor. In other words, the fact that my mother does match them at the expected 1st cousin level is proof enough in and of itself if we only had one cousin to test. We know our common ancestor is Cheryl and Don’s grandparents, who are my great-grandparents, Hiram Ferverda and Evaline Miller.
When I looked at Robin’s pedigree chart and saw that Robin descended from Philip Jacob Miller and wife Magdalena, I knew that this segment was a Miller side match, not a Ferverda match.
Therefore, matching with someone whose genealogy goes beyond the common ancestor of Cheryl, Don and me proves this line through 4 more generations. In other words, this DNA segment came through the following direct line to reach Me, Mother, Cheryl and Don.
- Philip Jacob Miller and Magdalena
- Daniel Miller
- David Miller
- John David Miller
- Evaline Louise Miller who married Hiram Ferverda
Clearly, we know from the earlier chart that my mother carried this DNA too, but even if we didn’t know that, she obviously had to have carried this segment or I would not carry it today.
So, even though in this example, our parents aren’t directly available for IBS testing and elimination, we can determine that anyone who matches both me and Cheryl or me and Don will have also matched mother on that segment, so we have, in essence, phased those people by triangulation, not by direct parental matching.
Step 8 – Triangulation Groups
What else does this match group tell us?
It tells us that anyone else who matches me and any one of our triangulation group on that segment also descends from the Miller descendant clan, one way or another.
Why do they have to match me AND one of the triangulation group members on that segment? Because I have two sides to my DNA, my Mom’s side and my Dad’s side. Matching me plus another person from the triangulation group proves which side the match is on – Mom’s or Dad’s.
We were able to phase to eliminate any identical by chance segments people on Mom’s side, so we know matches to both of us are valid.
On Dad’s side, there are some IBS by chance people (or segments) thrown in for good measure because I don’t have my Dad’s DNA to eliminate them out of the starting gate. Those IBS segments will have to be removed in time by not triangulating with proven triangulated groups they should triangulate with, if they were valid matches.
When you map matches on your chromosome spreadsheet, this is what you’re doing. Over time, you will be able to tell when you receive a new match by who they match and where they fall on your spreadsheet which ancestral line they descend from.
GedMatch also includes a triangulation utility. It’s a great tool, because it produces trios of people for your top 400 matches. The results are two kits that triangulate to the third person whose kit number you are matching against.
The output, below, shows you the chromosome number followed by the two kit numbers (obscured) that triangulate at this location, and then the start and end location followed by the matching cMs. The result is triangulation groups that “slide to the right.”
In the example above, all of the triangulation matches to me above the red arrow include either Mother, my Ferverda cousins or the Miller group that we discussed in the Just One Cousin article. In other words they are all related via a common ancestor.
You can tell a great deal about triangulation groups by who is, and isn’t in them using deductive reasoning. And once you’ve figured out the key to the group, you have the key to the entire group.
In this case, Mom is a member of the first triangulation group, so I know this group is from her side and not Dad’s side. Both Ferverda cousins are there, so I know it’s Mom’s Dad’s side of the family. The Miller cousins are there, so I know it’s the Miller side of Mom’s Dad’s side of the family.
Please also note that while this entire group triangulates within itself, that the group manages to slide right and the first triangulated group of 3 in the list may not overlap the DNA of the last triangulated group of 3. In fact, because you can see the start and end points, you can tell that these two triangulated groups don’t overlap. The multiple triangulation groups all do match some portion of the group above and below them (in this case,) and as a composite group, they slide to the right. Because each group overlaps with the group above and below them, they all connect together in a genetic chain. Because there is an entire group that are triangulated together, in multiple ways, we know that it is one entire group.
This allows me to map that entire segment on my Mom’s side of my DNA, from 10,369,154 to 41,685,667 to this group because it is contiguously connected to me, triangulated and unbroken. The most distant ancestor listed will vary based upon the known genealogy of the three people being triangulated For example, part of this segment, may come from Philip Jacob Miller himself, the line’s founder,, but another part could come from his son’s wife, who is also my ancestor. Therefore, the various pieces of this group segment may eventually be attributed to different ancestors from this particular line based upon the oldest common ancestor of the three people who have triangulated.
In our example above, the second group starts where the red arrow is pointing. I have absolutely no idea which ancestor this second group comes from – except – I know it does not come from my mother’s side because her kit number isn’t there.
Neither are any of my direct line Estes or Vannoy relatives, so it’s probably not through that line either. My Bolton cousins are also missing, so we’ve probably eliminated several possible lines, 3 of 4 great grandparents, based on who is NOT in the match group. See the value of testing both close and distant cousins? In this case, the family members not only have to test, they also have to upload their results to GedMatch.
Conversely, we could quickly identify at least a base group by the presence in the triangulation groups of at least one my known cousins or people with whom I’ve identified my common ancestor. Two from the same line would be even better!!!
Endogamy
The last thing I want to show you is an example of what an endogamous group looks like when triangulated.
This segment of chromosome 9 is an Acadian matching group to my Mom – and the list doesn’t stop here – this is just the size of the screen shot. These matches continue for pages.
How do I know this group is Acadian? In part, because this group also triangulates with my known Lore cousin who also descends from the same Acadian ancestor, Antoine Lore, son of Honore Lore and Marie Lafaille. Additionally, I’ve worked with some of these people and we have confirmed Honore Lore and Marie Lafaille as our common ancestor as well. In other cases, we’ve confirmed upstream ancestors.
Unfortunately, the Acadians are so intermarried that it’s very difficult to sort through the most distant genetic ancestor because there tend to be multiple most distant ancestors in everyone’s trees. There is a saying that if you’re related to one Acadian, you’re related to all Acadians and it’s the truth. Just ask my cousin Paul who I’m related to 137 different ways.
Matches to endogamous groups tend to have very, very long lists of matches, even triangulated, which means proven, matches.
Oh, and by the way, just for the record, this lengthy group includes some of my proven Acadian matches that were trimmed, meaning removed, from my match list when Ancestry did their big purge due to their new and improved phasing. So if there was ever any doubt that we did in fact lose at least some valid matches, the proof lies right here, in the triangulation of those exact same people at GedMatch
Summary
I hope this step by step article has helped take the Greek, or maybe the geek, out of matching. Once you think of it in a step by step logical basis, it makes a lot of sense and allows you to reasonably judge the quality of your matches.
The rule of thumb has been that larger matches tend to be “legitimate” and smaller matches are often discarded en masse because they might be problematic. However, we’ve seen situations where some larger matches may not be legitimate and some smaller matches clearly are. In essence, the 50% average seldom applies exactly and rules of thumb don’t apply in individuals situations either. Your situation is unique with every match and now you have tools and guidelines to help you through the matching maze.
And hey, since we made it to the end, I think we should celebrate with that beer!!!
______________________________________________________________
Disclosure
I receive a small contribution when you click on some of the links to vendors in my articles. This does NOT increase the price you pay but helps me to keep the lights on and this informational blog free for everyone. Please click on the links in the articles or to the vendors below if you are purchasing products or DNA testing.
Thank you so much.
DNA Purchases and Free Transfers
- Family Tree DNA
- MyHeritage DNA only
- MyHeritage DNA plus Health
- MyHeritage FREE DNA file upload
- AncestryDNA
- 23andMe Ancestry
- 23andMe Ancestry Plus Health
- LivingDNA
Genealogy Services
Genealogy Research
- Legacy Tree Genealogists for genealogy research


















Roberta,
We need to clarify once and for all, forever and always, the matching threshold for FTDNA. It is my understanding, according to both Michelle and Janine, that snp density is not factored in. Also, the minimum qualifying segment must be 5.5cM, and the remaining segments must add up to at least 22cM in total.
We really need to nail this down because there are too many conflicting reports. I know tgat you have sources!
SNP density has to be factored in because the lowest SNP number reported on any downloads are 500. I had a discussion with someone from FTDNA, not Janine or Michelle, about a month ago about match thresholds and the information in the article is what I was told. I think the clarification will have to come from within FTDNA but the article still applies regardless of what the matching threshold is.
Perhaps there is a minimum requirement on the largest qualifying segment (much like 23andMe)?
I appreciate the fact that you include endogamous segments in here. I have been recently exploring this possibility with some potential matches from Ireland. We have eight people triangulated on one segment (>10cM) on chromosome 9. Two are Irish by nationality and birth, and can document a common ancestor. The rest of us cannot find an Irish ancestor in our trees, nor have any recent (within 150 years) shared geographies. I share with the Irish folk 24cM! I was surmising that perhaps endogamy might be in play with the group. Very interesting abd thanks for the thoughtful post!
Rosario
Yes, that is the 7.7cM but there are some edge cases where there are other criteria which is why I said “about.” You know, sometimes these matches make perfect sense, and then there are those other times. Like your Irish. And then I think about how Anzick matches with some people at “current” levels and wonder how far back some matches really go.
I was told by another project administrator that Janine confirmed the 5.5cM longest block and I have seen LESS THAN 7.7cM longest block in my own matches. In fact I saw a 4.9cM, not sure how that got in there.
But it was said over & over again in the forums that the 7.7cM is what was mentioned on the ISOGG (wiki?) site.
I recently found a Estes-McDowell Link in Graves Co KY in 1850 census
1846 Sarah/Sally Estes married 1846 my ancestor Roland Via age 76
She was second wife of Roland Via who died in 1855.
So far have not found if Estes was maiden name or from previous marriage
“Those IBS segments will have to be removed in time by not triangulating with proven triangulated groups they should triangulate with, if they were valid matches.”
I actually understood that sentence.
Great article. This one will be at the top of my list instead of the “No Zipper” one. “No Zipper” is going down to number two, unless you edit the “No Zipper” post and add a picture of a kitten. Then I won’t know what to do.
A kitten. What a great idea….
Roberta, how do you decide when to update your master spreadsheet?
Does it contain the DNA matching segments of all of your matches as well as those of your relatives (i.e., you manage their kits or they email you their Chromosome Browser CSVs, etc.)?
I don’t update it nearly as often as I should. You can download just a few matches at a time through the chr browser. Only matches to me and my Mom are in the master spreadsheet. When I’m working on a project with particular cousins, I create something different for that project, like the Vannoy project for example. I don’t know that this is the best way, but it’s the only way I can stay unconfused.
That’s perfect. I dont mind having a 50,000 line spreadsheet. It’s the not getting confused part that Ive had a hard time with. I just need to make sure i keep good notes. Thanks.
Roberta,
I always find your articles very informative but often have a great deal of difficulty in figuring out how to apply them to my situation which is an endogamous group. Just working with downloads is difficult because they are so large that getting to a particular segment you are interested in means paging though a lot of lines just to get there. Maybe there are some spreadsheet tricks I just don’t know about (I use OpenOffice Calc). Also, I’m not sure about comparison by alleles vs comparisons by segment. When you match on a segment does it mean that you are matching on both alleles or just one of the possible two?
I am further handicapped by knowing very few family surnames other than my grandparents and that’s only on my maternal side. I don’t know anything about the parents of my paternal grandparents. All of my known antecedents are from Russia and immigrated in the late 19th or early 20th century. While some of my matches have family trees going back much farther than mine we can not identify any common surnames, most likely I think because they came through a female ancestor whose maiden name has been lost to time. There is no one left alive to test from the generation preceding mine and there aren’t many of us left to get tested.
So in short, my biggest problem is just where should I start digging into this mess. All family tests results are now are on FTDNA although a couple, including mine were transferred from Ancestry. My sister was tested on FTDNA and I notice she has SNPs that I don’t. We match on over 2500 cMs but she seems to have more higher level cousin matches than I do even on the same people. Is that because of the extra SNPs or is it an indication she is bringing in a lot more DNA from one of my parents than I am? Can In Common and Not in Common matches tell me something? What about the Matrix and who would I want to put in a Matrix?
Any advice would be greatly appreciated. Feel free to respond by email if you don’t think anyone else here would be interested.
You know, the only place you can start is with what you know. Test any cousin or anyone related you can find. That and surnames may be all you have to begin. It sounds like you are from an entirely endogamous group. I would like very much to see Israel or someone else with an entirely endogamous family to work with this type of situation. As for your sister, yet, she has different DNA than you do – 50% different.
There may be spreadsheet tricks, but I do mine by hand. It has about 20,000 lines now with both me and my Mom’s in the same file.
June, which company did you have relatives test at? The only difference I saw in SNPs was when I compared my mother’s V4 23andme results to my V3 results. My brother also tested no V3 but his differences was not that obvious with the exception of the ancestry portion.
With endogamy you will want to concentrate on the longest block for your closest matches. Anything below 20cM can be nearly impossible.
I originally tested at Ancestry and transferred my data to FTDNA. My sister tested directly at FTDNA. When I went to download raw data (I have access to her kit) the first thing I noticed was that there were two different downloads available and since there was no explanation on the page about how they differed I choose B37 which I think is the “advanced” form, whatever that means. Just looking at the first page of chromosome 1 data I found that she had 3 more SNPs listed than I did matching by location in the order listed. We matched on all but two of the ones we both had and one of those was one where I showed a 00 rather than an allele pair.
BTW, thanks for the tip about concentrating on 20cM and above. I don’t have many so that concentrates here I need to focus on. Apparently FTDNA thinks total cM is more important that longest cM as the default order in which my matches are listed displays longer total cM above shorter ones with a shorter total but a longer longest segment.
Awesome article! I have a spreadsheet similar to yours but with different colors – light green for Dad’s side and light orange for Mom’s side. I have also across IBS or “false” matches at FTDNA while doing comparisons but they were in the 6 cM range.
Pingback: DNA Saturday | Diggin' Up Graves
Thank you for this article. Triangulation is a great way to develop a picture of relatedness using Autosomal DNA. Our Amerindian Ancestry project finds that there is high repeatability for Autosomal DNA matches, by way of triangulation, among parents and their offspring, siblings, and others who share recent, known ancestry connections.
No question. It is very useful to have the DNA test results of at least one parent when proofing your Autosomal DNA matches (and your paper trail genealogy).
Good article, Roberta. Thank you. Two comments.
1. In step six, you answer the IBD question (#2) “yes” a,d then ask if the match is identical by population. You might want to explain why it is useful to ask that after yu already know you have IBD. (I think I know, but I’d rather hear it from you.)
2. I worry about the “slide right” group. Not that I don’t trust what you did with your own known people. It’s just so easy to go wrong.
Hi Israel. 1. Unfortunately, our DNA does not come with a timestamp on it. Don’t we wish it did. So once we’ve determined that it is identical by descent, it could be from a common ancestor a generation ago, or maybe Anzick, 12,000 years ago. Yes, those are both extreme ends of the spectrum, but they are clearly both possible. Some IBD you will be able to prove by genealogy, and the rest may be too far back, which are identical by population. These can certainly be hinted at when you have multiple ancestral lines that match to the same segments from the same side of your family. 2. I agree with you about the slide group. I was very careful in my illustration to select only groups that had only people I knew to be in a cluster, in the group. Many groups intermix your ancestors from both sides of your family because the DNA addresses are the same, of course. In time, if you have a parent’s DNA, you would be able to get two triangulated groups, but for not, no. I debated about using this illustration at all, but I think it does illustrate, if you’re careful, and not endogamous on both sides, how they can potentially be used. I know that doesn’t help you much:)
Thank you Roberta . This one is a keeper ,will be in my three ring binder for future reference.
Hi Roberta
Another great post, I just need to ask if the following scenario has cut any corners for me.
I tested Ftdna and 23andme ( v3) and my son 23andme ( v4). in 23andme he has phased me and received a “split view” which also tell him which matches he has which are on the paternal side or maternal side.
He also phased his gedmatch account and has 2 other gedmatch accounts ( kit endings in P1 and M1 )
Whats the best way forward for me from here, the Kitty Cooper tool or the gedmatch.
regards
victor
I don’t understand the question. To the best of my knowledge, Kitty’s tools and the GedMatch tools don’t compete or perform the same functions, and they are both free or contribution bases, so very minimal. I’d try them all.
Roberta
As someone said above – this article is a keeper.
Inspired by it, I applied some of the principles to one of my “cousins” at 23andMe. This person according to them is 3rd to 5th cousin – We share 0,72% on two segments total 54 cM. One of the interesting thing is that the longest of the two, 37,6 cM is on a X chromosome. So from the start I suspect that 3rd to 5th is probably very optimistic.
Because my two X chromosomes look very different in terms of ethnicity and my son also matches, I know that the link is most likely through my paternal grandmother. Fortunately my “cousin” also has an extensive tree. To find the common ancestor I look at all possibilities, knowing that men only pass X to their daughters, in my tree, then look for the names to be present in her tree. My first candidate does not work, because while she is also related, her line involves two males in a row.
Finally last night – got it. Two potential sisters, Genevieve and Marguerite Albert – I have the parents – she doesn’t, but I confirm on someone else’s tree – Pierre Albert and Louise Therese Grondin married in 1702 in Riviere Ouelle, Kamouraska, Quebec. This is 10 generations!
But it is the X chromosome and we all know “rules” don’t apply.
However, what bothers me most is that while my son matches and his matching segment starts at exactly the same spot, it is only 10 cM long? How could that be?
We are also working with an endogamous population. I think that maybe I am also linked through my mother. So back to the search and yes! I find another common ancestor that fits. Interestingly enough, for my cousin (at 10 generations) she links up fairly early, so the line is the same as the other one. For me, it is my direct matrilinear line. The common ancestor this time is Anne Gautier born in Port-Royal, Acadie in 1722. This time it is 9 generations for me.
So we are related twice, once at 10 generations, the other at 9 generations. The other thing that is amazing is that the two segments that lie on my two X chromosomes are adjacent! In my cousin’s case, it less surprising since they are carried by the same persons through 6 generations.
In summary, I think this is a good example of applied autosomal genealogy. It is essential to have a well documented tree to be able to find common ancestors, because they can go very far back.
That’s a great application of the technology. Ironically, I’ve been working with a researcher on a segment of X too, and it’s not playing nice either.
Roberta,
I have been Demystified. Seriously. It’s really all falling into place. My spreadsheet, the missing pieces, the “Why didn’t I see this when I was looking for the connection with this person six months ago? It was right there all along!” thoughts.
Maybe it’s the case of just having to get to that point of providence where the background, the brain capacity, and the new information all fall into place.
I have slept, but most of my waking hours in the past two days have been spent working on this. You are probably one of the few people who would understand how a spreadsheet can give so much excitement.
Thank you for continuing to respond to our needs (and mine) often before we even know what they are. The genealogical community owes you a debt of gratitude, as do I.
Elizabeth Ballard
Pingback: A Study Utilizing Small Segment Matching | DNAeXplained – Genetic Genealogy
Pingback: Friday Finds – 01/23/15
Pingback: Samuel Muncy (1761/1768-1839), Who’s Your Daddy, Your Mamma, and Your Kids?, 52 Ancestors #56 | DNAeXplained – Genetic Genealogy
Pingback: Secondary Genealogical and Genetic Lines | DNAeXplained – Genetic Genealogy
Pingback: DNAeXplain Archives – Intermediate DNA Articles | DNAeXplained – Genetic Genealogy
Pingback: The Best and Worst of 2015 – Genetic Genealogy Year in Review | DNAeXplained – Genetic Genealogy
Pingback: Concepts – Why Genetic Genealogy and Triangulation? | DNAeXplained – Genetic Genealogy