This is Part 5 of a series.
Part 1 was “The Autosomal Me – Unraveling Minority Admixture” and Part 2 was “The Autosomal Me – The Ancestors Speak.” Part 1 discussed the technique we are going to use to unravel minority ancestry, and why it works. Part two gave an example of the power of fragmented chromosomal mapping and the beauty of the results. Part 3, “The Autosomal Me – Who Am I?,” reviewed using our pedigree charts to gauge expected results and how autosomal results are put into population buckets. Part 4, “The Autosomal Me – Testing Company Results,” shows what to expect from all of the major testing companies, past and present, along with Dr. Doug McDonald’s analysis.
In this segment, Part 5, we’re going to look at various third party tools and what they can do for our search for minority admixture. We will use the download files from either 23andMe and Family Tree DNA and utilize third party tools to analyze the raw data. We’ll see how third party developers put those puzzle pieces together, if the results are consistent and what they tell us.
The Weeds
When dealing with testing companies, particularly any individual source (as opposed to multiple testing company results, as I have done), minority admixture, especially less than 1% may not be successfully recognized. One percent equates to between 6 and 7 generations or about to the 1800 threshold in time. However the history of both African and Native admixture in colonial America goes back another 200 years to the Jamestown era.
The social history in the US means that there are many people looking for this admixed heritage as long ago as 1609 when Jamestown was established and the first European/Native marriages took place (although there were “blonde Indians” reported by Jamestown settlers). In round numbers, that’s about 400 years or between 13 and 16 generations. Of course, a minority ancestor drops below the 1% threshold between 7 and 8 generations (with the first generation being the person tested) and by the time you get to the 12th generation, you’re at .048%. At this level, Bennett Greenspan says we’re “rooting around in the weeds,” and he’s right.
However, rooting around in the weeds for those dreaded IBS (Identical by State) segments in genealogy is exactly what we need when looking for small amounts of minority admixture. What’s an IBS segment you ask? It’s a segment that is typically too small to be counted as an IBD, or identical by descent, segment. IBS means that you’re from a common population if you match someone with a very small segment, not necessarily that you share a common ancestor within the past several generations. But how to you tell if a small segment is IBS or IBD?
There is no absolute line in the sand, but often segments smaller than 7cM (centimorgans) or 700 SNPS (some say 5cM and 500 SNPs) fall into the IBS category. This has caused some researchers to discard all segments of this size because they can’t tell the difference. That’s unfortunate, because clearly some of these segments are IBD and the IBS segments can be useful too.
When looking for minority admixture in two people, both of them having these small segments in the same location can provide meaningful information, and can confirm minority heritage. Said another way, if two people have less than 1% Native heritage, both share a common ancestor, and both carry part of their “less than 1%” on the same segment….one might say it’s not likely to be coincidence. Identifying the common segments of your common ancestor can lead to identifying the specific family line those segments came from, especially if you match others as well. This is in essence what Minority Admixture Mapping, or MAP, does. It uses these techniques to look for patterns in these small fragmented pieces that, when taken together, indicate minority heritage. Having said that, some IBS segments will indeed, be simply that, because you share the same base population, but some will be IBD, or more current in time. With the MAP technique, we’re sorting through ways to utilize these small segments, whether they are IBS or IBD.
Using the tools, MDLP, Eurogenes, Dodecad and HarrappaWorld at GedMatch allows us to “root around in the weeds,” to quote Bennett, and find those all-important small IBS/IBD segments that connect us to a particular ethnicity and ultimately, to other relatives who carry these same segments in the same locations.
In general, using these this type of DNA is called BGA, or Biogeographical Ancestry where we use SNPs of autosomal DNA called AIMs, Ancestry Informative Markers. A SNP is a Single Nucleotide Polymorphism, or a mutation that happened in one specific location on a gene. AIMs are generally SNPs, not clusters of markers, found at different frequencies in different populations. We combine all we know about them scientifically with information about population frequencies and then draw inferences about where our ancestors came from based on that information. So a SNP that is useful in determining ancestry is called an AIM.
These SNPs, or AIMs, are the foundation for these BGA tools that we will be using to sort through small segments of minority admixture. So this is a building block process. Scientists identify SNPs found in different populations at different frequencies and identify them as such, then scientists and genetic genealogists create BGA tools that use and combine SNPs/AIMs to suggest populations and ethnicities for those who carry them. Using these tools, majority ancestry is easy to discern. We’re going to use those tools to look at groups of SNPs/AIMs clustered in small, fragmented IBS or IBD segments to do Minority Admixture Mapping (MAP) to confirm our minority admixture and to identify our minority admixed lines, families and perhaps even (in time) our original minority ancestor.
I bet you thought I couldn’t fit all of those acronyms in one paragraph, but I did:) It is a bit like alphabet soup, but when you understand that this is a building process, it’s much easier to grasp as a whole.
Having at least one parents DNA makes this process much easier, because you can immediately tell if your other parent, by inferrence or process of elimination, has contributed any of the minority ancestry, or if it’s all on one side of the tree. Of course, that’s assuming your parents aren’t related to each other. There’s a test for that too at GedMatch. If you don’t have one parent available, you can “make do” with aunts, uncles and cousins, but it’s a much more tedious process.
Third Party Tools
To use any of these BGA tools, you’ll need to download your results from either 23andMe, Family Tree DNA or National Geographic. Currently at GedMatch, the only supported formats are 23andMe or Family Tree DNA, because the National Geographic test is so new. I used my Family Finder (Illumina Build 36) raw data file.
To download your results from 23andMe, sign on to your account, then click on this link and it will take you to the area to download your results.
https://www.23andme.com/you/explorer/
Save the file and do not open it as the act of opening it sometimes causes corruption and you will have a hard time uploading the file. If the upload fails, download a new copy and start over. If you have an older copy on your computer, it’s always a good idea to use a fresh copy to incorporate any changes made by the vendor since your last file download.
To download your results from Family Tree DNA, sign on to your personal page, click on the Family Finder tab and then on “Download Raw Data.” As I write this, Family Tree DNA is in the midst of a conversion from Build 36 to Build 37 for their autosomal files (in order to facilitate the integration of 23andMe results), so you may need to be a bit patient while this process completes. Files may not be available for download at some points. You certainly don’t want to mix comparisons, meaning using one build 36 and one build 37 file for comparison.
If you’re following this process yourself with your own data, please read all the way through this posting before starting your own processing.
Now, let’s look at the third party tools.
Stanford University
This tool is available at Stanford University. Scientists have collaborated to provide this service and I think it’s quite interesting. This tool is not compatible with any browser except Chrome and it requires a download of your autosomal data in a .txt file. If it can’t load your file, the loading task simply never completes. For me, that meant it wasn’t a .txt file I was trying to load.
Load your file and choose Ancestry, then Paintings, then Hap Map 3 (experimental), then Paint my Chromosomes.
Their legend, above, translates to the regions, below.
ASW – African ancestry in Southwest USA
CHD – Chinese in Metropolitan Denver, Colorado
GIH – Gujarati Indians in Houston, Texas
LWK – Luhya in Webuye, Kenya
MEX – Mexican Ancestry in Los Angeles
TSI – Toscani in Italia
Unfortunately, this isn’t terribly useful. Hap Map 3 utilizes additional regions, including Utah, but this tool doesn’t seem to be mapping them, so my closest match region is Italy, which is midleading since none of my family was from Italy. Hap Map 2 is also an option which does include the Utah population, but it’s not as up to date otherwise as Hap Map 3.
David Pike has figured out how to tweak these settings some. You can read about it at this link: https://www.23andme.com/you/community/thread/8062/. David’s posting on June 20th shows what he did. However, compared to the other tools available, I find this a poor choice and did not spend a lot of time trying to work with it.
However, a second feature that they provide is fun.
Stanford provides a Neanderthal tool that’s a little different than the Nat Geo or 23andMe ones. Click on Explore, Neanderthal, Look Up Exercise. Then enter your primary ethnicity and click on Look Up Exercise again.
Of a possible 84 Neanderthal alleles, I have 9, partially displayed below.
GedMatch
www.Gedmatch.com is a complimentary (voluntary contribution) site created by two genetic genealogists that includes several autosomal analysis tools. One of the areas of this site is “Admix Tools.” On that page one finds several private or proprietary tools, some written by genetic genealogists, some by researchers, and all free. Let’s take a look at each one and their results. If you want to see any of the results more closely than the photos here allow, you can run each of the comparisons using kit F6656 (mine) as the first kit and kit F9141 (my mother) as the second kit.
Each of these tools offers the same functionality, as follows.
We will be utilizing 4 of these functions for each tool.
- Admixture Proportions
- Admixture Proportions by Chromosome
- Chromosome Painting
- Paint Differences between 2 kits, 1 chromosome
We select from the tools as follows:
Let’s take a look at what the tools provide.
MDLP World 22
The MDLP software is sponsored by two genetic genealogists. You can read more about the project at http://magnusducatus.blogspot.com/ and http://magnusducatus.blogspot.com/2012/09/behind-curtains-mdlp-world-22-showcase.html.
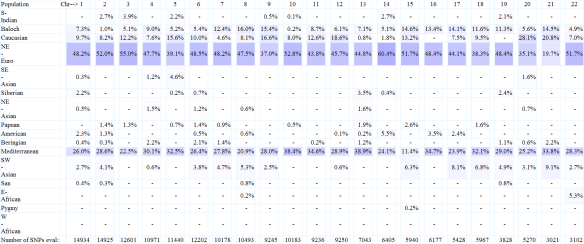
MDLP shows several populations. I was interested to see if my mother also shared the African percentage. Interestingly, mother does have a South African segment, but it’s .12, so less than mine. Therefore, I would have obtained part of my African heritage from my father. She also has three different categories of Native American heritage, compared to my one. She carried a total of 1.92% and I carry .58%. Otherwise, our results are very similar.
The next feature is ethnicity mapping by chromosome. While the display is too large to see well it’s interesting to note that indeed, both Native American and African were detected on several chromosomes, not just on chromosomes 1 and 2 as reported by 23andMe and Dr. McDonald. Note that DeCode Genetics showed “East Asian” admixture on several chromosomes.
Here’s a portion of the above chart that you can actually see. The highlighted blue regions are your major ethnic regions.
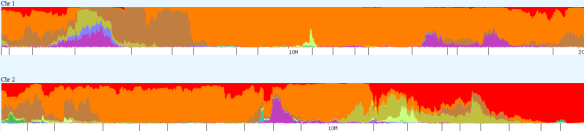
Another feature is chromosome painting, shown below. This shows the first part of my chromosomes 1 and 2 painted by ethnic/regional breakdown. The legend for each tool is different and above their graph.
These tools also provide the ability to compare one chromosome between two people. On the graph below, my chromosome 1 is on the top, and my mother’s is second, with the third band being our common painting. The black represents non-shared regions, meaning those contributed to me by my father. Unfortunately, North American Native American is dark grey, sometimes difficult to distinguish from black.
The graph below shows that while I do share a large piece of Chromosome 1’s Native region (about 160-180mb) with my mother, there are also segments, 169-170 for example, where I have Native genes that she does not, indicating Native heritage in this location from my father’s side.
Eurogenes K9
Eurogenes was created by another genetic genealogist. You can read more about it at http://bga101.blogspot.com.au/2012/04/eurogenes-admixture-utilities-at.html.
Eurogenes calls me primarily North European with .67 Native American and no African in the percentages above, but below, on the individual chromosomes, some African does show, although not on as many chromosomes as MDLP.
In the charts above and below, you can see that Eurogenes detected small amounts of African along with Native American.
Notice that at about 10mb on chromosome 1, on the graph below in the top band, that the North American Indian (yellow) and the South Asian (red) are imbedded with each other. These appear again together at the beginning of chromosome 2, shown as the second band. This hints at how and why it’s sometimes so difficult to determine and filter Native American from Asian. There is no line in the sand, there is a continuum between populations, the only differentiator being 10,000 to 15,000 years spent apart in which time, they, hopefully, developed enough differentiating mutations that we can tell them apart.
On the chart below, the top band shows the chromosome painting of my chromosome, and the second band shows the chromosome 1 Native American segment (about 160-180 mb) of my mother with the third band showing both matching and non-matching regions, painted black. Looking at the segment of chromosome 1, in the graph below, characerized as Native, we can see in mine, top row, that this is categorized as Native American (yellow), but some of the same regions below, in Moms are categorized as South Asian (red), causing a technical non-match, when in reality, It’s likely a categorization issue, not a genetic mismatch. In future analysis, we’ll be using two methods of comparison, one called “Strong Native” that only matches Native to Native and another, the “Blended Asian” method that allows for grouping of similar ancestral types that together likely indicate a Native heritage.
Dodecad V3
Dodecad was created by an anthropologist. You can read more about it at http://dodecad.blogspot.com/ and http://dodecad.blogspot.com/2011/06/design-of-dodecad-v3.html.
Dodecad, unfortunately, does not subdivide into Native American, so the Native will show here as some form of Asian. Northwest Africa shows in the percentages above, but more detailed African heritage shows in the chromosome detail below in regions not shown above.
Above, my chromosome painting for the first part of chromosomes 1 and 2.
Below, the comparison showing the Native segments from about 160-180mb. My Native segment (top) compared to mother’s (middle) with the comparison of the two on the bottom for chromosome 1.
HarappaWorld
HarappaWorld divides results into fewer population groups and is focused on Asia. You can read more about it at http://www.harappadna.org/2012/05/diy-harappaworld/.
In HarrappaWorld, Beringian and American appear to be equivalent to Native American. Like Dodecad and Eurogenes, African does not show in the total percentages, but does on the individual chromosome analysis, although in smaller percentages with this application.
Chromosome painting of my chromosomes 1 and 2 are shown below.
The graphs below show the Native region comparison of chromosome 1 between me, top row, mother, middle row, and the third graph showing the common areas, with black representing areas where there is no match.
For each of these tools and their results, we’ll do further analysis in a future segment of this series.
Tools Summary
Now that we’ve looked at these individual tools, and building on the Test Results Chart created in Parts 3 and 4, let’s compare and see what information these tools add.
Test Results Chart Including Third Party Tools
| Test/Company | European | Asian | Native | African | Unknown |
| Pedigree Analysis |
75% |
0 |
~1% |
0 |
24% |
| Testing Companies | |||||
| Family Tree DNA – Original |
100%[1] |
0 |
0 |
0 |
|
| deCodeme |
92% |
5% |
Inferred[2] |
3% |
|
| deCodeme – X |
91% |
6% |
Inferred |
3% |
|
| Dr. McDonald |
97-99% |
1-3% |
0.5% |
0 |
|
| 23andMe – Original |
99% |
1% |
Inferred[3] |
0 |
0 |
| 23andMe – 2012 – Standard |
99.2%[4] |
0 |
.5% |
0 |
.3% |
| 23andMe – 2012 – Conservative |
98.7%[5] |
0 |
.3% |
0 |
1% |
| 23andMe – 2012 – Speculative |
99.3%[6] |
0 |
.5% |
0 |
.2% |
| Family Tree DNA – 2012 |
100%[7] |
||||
| Geno 2.0 |
79%[8] |
18% |
0 |
0 |
0 |
| Ancestry |
92%[9] |
0 |
0 |
0 |
8% |
| Third Party Tools | |||||
| MDLP |
86.68% |
12.55% |
.58% |
.17% |
0 |
| Eurogenes |
94.83% |
4.5% |
.67% |
0 |
0 |
| Dodecad |
85.47% |
13.43% |
Inferred |
1.09% |
0 |
| HarrappaWorld |
86.56% |
12.80% |
.65% |
0 |
0 |
Of the various chromosomes, the breakdown is as follows. Dodecad does not break the categories in a comparable fashion to these other 3 tools, so their results are omitted in the following chart. Please note that how geographies are categorized can make a significant difference.
Minority by Chromosome Chart
| Tool/Chr | MDLP Native | Eurogenes Native | Harrappa Native | MDLP African | Eurogenes African | Harrappa African |
| 1 | Y | Y | Y | N | N | N |
| 2 | Y | Y | Y | Y | Y | N |
| 3 | N | N | N | Y | Y | N |
| 4 | Y | N | Y | N | N | N |
| 5 | N | N | N | N | N | N |
| 6 | Y | Y | Y | Y | N | N |
| 7 | N | N | Y | N | N | N |
| 8 | Y | Y | Y | Y | N | Y |
| 9 | Y | N | N | Y | N | N |
| 10 | Y | N | N | Y | N | N |
| 11 | Y | N | Y | Y | N | N |
| 12 | Y | N | Y | N | N | N |
| 13 | Y | N | Y | N | N | N |
| 14 | Y | Y | Y | Y | N | N |
| 15 | Y | N | N | N | N | Y |
| 16 | Y | Y | Y | Y | N | N |
| 17 | Y | Y | Y | N | N | N |
| 18 | N | N | N | N | N | N |
| 19 | Y | Y | Y | Y | N | N |
| 20 | Y | Y | Y | Y | N | N |
| 21 | Y | N | Y | N | N | N |
| 22 | N | N | N | Y | Y | N |
From these various tools, it’s obvious that I do have some Native admixture, probably about 1%, and it’s from both parents. I also have some African, but it looks to be an even smaller proportion that Native American.
Join me for Part 6 of this series, where we look at how to analyze and use this information.
[1] 71.5% western European, 28.4% Northeastern European
[2] Inferred that Asian is actually Native in an American with no history of Asian ancestry.
[3] No category, inferred.
[4] 78.6% Northern European, 1.8% Southern European, 18.7% Nonspecific European
[5] 54.6% Northern European, .3% Southern European, 43% Nonspecific European
[6] 91.7% Northern European, 3% Southern European, 3.3% Nonspecific European
[7] 75.18% West Europe (French and Orcadian), 24.82 Europe (Romanian, Russian, Tuscan and Finnish). Note that my mother’s results are almost identical except the Finnish is missing from hers.
[8] 43% North Europe and 36% Mediterranean
[9] 80% British, 12% Scandinavian
______________________________________________________________
Disclosure
I receive a small contribution when you click on some of the links to vendors in my articles. This does NOT increase the price you pay but helps me to keep the lights on and this informational blog free for everyone. Please click on the links in the articles or to the vendors below if you are purchasing products or DNA testing.
Thank you so much.
DNA Purchases and Free Transfers
- Family Tree DNA
- MyHeritage DNA only
- MyHeritage DNA plus Health
- MyHeritage FREE DNA file upload
- AncestryDNA
- 23andMe Ancestry
- 23andMe Ancestry Plus Health
- LivingDNA
Genealogy Services
Genealogy Research
- Legacy Tree Genealogists for genealogy research































Pingback: Part 5 of The Autosomal Me Series | Native Heritage Project
Roberta, I do not know where to begin. You are awesome!! You are a scholar! What a labor of love to share with your following. Why aren’t you doing webinars on just this blog?? You could make a mint.
Thank you so much. I have considered webinar and may do more in the future. I agree that this would be a good topic.
I second the request for webinars! I prefer setting aside time to watch and listen. Trying to follow all the blogs or sitting down and reading is OK, but watching it would be more effective for me.
I liked you report. I did my husband from 23 and me to GED match it was interesting.I wish that mine could have been done also. I uploaded mine first and realized that it was mine with some of his hapo group and got to load his right finally. It was neat checking out all the graphs. Thanks Jo
Well, now that I have tested with 23andME and NatGeo, I still don’t know what to do. I don’t think I know how to get all those programs tweaked on my computer. I had problems with some websites that were sending me things that I didn’t want to receive. That’s not to say that 23andME sent them, just my looking around. I even got on to a website from Russia and have received invitations to collaborate with people who are not even in my haplogroup. I have to be careful how much junk I have coming into me.
One of the sub-selections under the Dodecad tool of GEDmatch is World9, which according to the developer was designed to address Native American percentages more accurately. It appears to be a better tool than V3. Have you compared it with the others you mention?
No Mark, I haven’t. When I started doing this, I don’t think it was available. I would certaily suggest that anyone undertaking this path take a look at that.
I have been experimenting with the various ad mixtures at gedmatch and trying to ‘wrap my mind around’ all the information. Your blogs help a lot.I find it amazing to look at the chromosomes and see traces of distant ancestors. I have a small amount of Native American from my mom( my fathers parent’s were from Italy). My mom and her parents have all passed. I just wish I could get more people from her family to test. I’d love to pin down whether the Native American is all from her paternal side ( some ancestors from Spain/Mexico most probably situation) or if some of the Native American from her maternal side ( colonial american roots with a rumor of Native American). Both her maternal and paternal side have dead ends so anything is still possible.
Hi Roberta,
Thank you very much for ‘The Autosomal Me’ series, I am finding it very useful and educational. I have looked at a few of the admixture tests from Gedmatch but I need a bit of help interpreting them. My paper trail research for my Dad’s 32 great great grandparents is 59% Irish, 38% English and 3% unknown (possibly Irish but family oral history suggests otherwise). The admixture tests at Gedmatch with the Oracle test are bringing up some unexpected primary and secondary populations. As an example, Oracle for Dodecad K12b the top populations are 92.1% Cornwall (1000Genomes) and 7.9% Iranians (Behar). I’d like to know whether Dad’s admixture results are typical of people with Irish/English known ancestry or whether I actually have something interesting to pursue here. Can you suggest any place to see typical admixture percentages for people of predominantly English/Irish origin? Thank you again for your great blog.
Probably the best reference I’ve seen for admixture of typical populations is the National Geographic Geno 2.0 autosomal results explanations of populations. If you have not tested there, I have some of them posted on my blog at http://dna-explained.com/category/genographic-project/ under the Genographic category, but particularly under this one: http://dna-explained.com/2012/12/12/geno-2-0-results-kicking-the-tires/
Thanks Roberta, I will try those posts about Geno 2.0.
Re: the colors MDLP uses, in addition to North-Amerind’s charcoal grey being very close to black, MDLP World 22 uses the same color for some of the groups. I used a digital color meter to determine the following are identical: Pygmy/Sub-Saharian, West-Asian/East-South-Asian, North-European-Mesolithic/Near_East, Indo-Tibetan/Melanesian, Mesoamerican/Paleo-Siberian, Arctic-Amerind/Austronesian. What’s the use of MDLP’s chromosome painting if it makes no distinction between two highly divergent groups like Austronesian and Arctic-Amerind?
Well, I’m glad to know it’s not just me. I had fits with these.
Other good tools to try are the DIYDodecad programs such as Globe13 which can be downloaded to the computer from Dienekes Dodecad blog site, and best of all, they are free. One can dig deeper into every DNA segment using these programs and can even plot out one ancestry, or two ancestries, or more on a graph. These graphs enable one to see minority peaks better than the chromosomal paintings at Gedmatch since sometimes the minority peaks are hiding behind the big peaks in the paintings.
I tried using the admixture tools on gedmatch to confirm the sliver of African DNA (0.1%) in my grandma’s 23and me results. All four tools show the African DNA ib the graphs, but only one lusted it in the percentage breakdowns. Do you kbow why this would happen? I have screenshots on my blog at http://brandtgibson.blogspot.com.
Thanks,
Brandt
I would suggest that you contact the developers. Their e-mails are listed on the GedMatch site. My guess would be because they are less than 1%, but if that’s not the case, I don’t know.
I don’t understand how to read MDLP World22 Single and Mixed Mode Population Sharing. What does the “distance” mean?
I don’t know off the top of my head, and I’m not in a position or place where I can work with that data right now. There is a link to contact the developer. Maybe you should ask them. Sorry I can’t be of more help right now.
Pingback: DNAeXplain Archives – Intermediate DNA Articles | DNAeXplained – Genetic Genealogy
I’m reading some old article from your archives list. I don’t recall reading this one.
First, thanks for posting this one, it really help to compare with someone else result. So, it appear I have more Amerid blood than expect, about 1.67%, while my pedigree hint to 0.1% Abenakis, 0.1% Ojibwe and 0.1% Mi’kmak… I better do my pedigree chart with an erasable pen.
Hi-
On Eurogenes K13, I get 0.41% Amerindian ancestry. Then, on Eurogenes K9, I get 0.43% Native American. Furthermore, on Eurogenes Hunter_Gatherer vs. Farmer, I get 0.4% South American Hunter Gatherer. Additionally, on puntDNAL K10, I get some Amerindian too.
Do you think that this Native ancestry is real?
It’s very small. I really can’t say. Remember that Germanic people also get as high as 3%.
Pingback: Concepts – Managing Autosomal DNA Matches – Step 2 – Updating Match Spreadsheets, Bucketed Family Finder Matches and Pileups | DNAeXplained – Genetic Genealogy
Thanks for explaining this subject so well! I had two questions though which I haven’t been able to find answers to – I don’t know if you have time to answer. In the chromosome paintings, I see you can have a number of different population groups in the same centimorgan (e.g. all of the peaks and valleys) – is this because there are hundreds of different SNPs in each of the centimorgan regions mapped, so the paintings are showing the population group breakdown of the smaller units that make up the region, or is it just showing the percentage likelihood that any particular region belongs to one (but not all) of the different population groups listed? Also, if you are showing a region that is more than 50% coloured for one population group, does this mean that you must have inherited the DNA from that population group (if accurate) from both parents, or does recombination work so that some regions would contain more or less than 50% from either parents (with the total of all regions on the chromosome being 50% from each parent)? These also may be silly questions and I may not be getting this at all but thanks for your efforts!
Apologies for the second question – of course each and every region would need to be 50% from each parent. So if a region shows as more than 50% for a population group then both parents would need to have contributed – correct?
Yes, assuming the estimate is accurate.
Thanks for the very quick reply! This can be helpful in filtering out noise where it is highly unlikely a population group would be contributed by both parents.
The Sanford University DNA snippets about your local autosomal DNA results is now here:
http://genotation.stanford.edu/