This is Part 7 of a multi-part series.
Part 1 was “The Autosomal Me – Unraveling Minority Admixture” and Part 2 was “The Autosomal Me – The Ancestors Speak.” Part 1 discussed the technique we are going to use to unravel minority ancestry, and why it works. Part two gave an example of the power of fragmented chromosomal mapping and the beauty of the results.
Part 3, “The Autosomal Me – Who Am I?,” reviewed using our pedigree charts to gauge expected results and how autosomal results are put into population buckets. Part 4, “The Autosomal Me – Testing Company Results,” shows what to expect from all of the major testing companies, past and present, along with Dr. Doug McDonald’s analysis. In Part 5, “The Autosomal Me – Rooting Around in the Weeds Using Third Party Tools,” we looked at 5 different third party tools and what they can tell us about our minority admixture that is not reported by the major testing companies because the segments are too small and fragmented.
In Part 6, “The Autosomal Me – DNA Analysis – Splitting Up” we began the analysis part of the data we’ve been gathering. We looked at how to determine whether minority admixture on specific chromosomes came from which parent.
Part 7 – “The Autosomal Me – Start, Stop, Go – Identifying Native Chromosomal Segments”, takes a deeper dive and focusing on the two chromosomes with proven Native heritage, begins by comparing those chromosome segments using the 4 GedMatch admixture tools. In addition, we’ll be extracting Native segment chromosomal start and stop addresses that we’ll be using in a future segment.
Using Doug McDonald’s tool and the 23andMe results, we can begin with the following two Native segments, one each on chromosome 1 and 2. These will be our reference points, because according to both sources, these are the largest and most pronounced Native segments, the strongest indicators, so they will be our best yardsticks.
| Chromosome 1 | Chromosome 2 | |
| 23andMe |
165,658,091 to 175,711,116 |
86,316,174 to 103,145,426 |
| McDonald |
165,000,000 to 180,000,000 |
90,000,000 to 105,000,000 |
On all of these admixture graphs, my results are shown first, then mother’s, then the comparison between the two where the colored regions show common ancestry and the black shows nonmatching segments – in other words those contributed by my father.
Please note that Native contribution in this analysis is being evaluated by a combination of geographies. In some cases, one individual will show as “Native” meaning in the case of MDLP “North Amerindian” and the parent (or child) will show as something similar, like “Actic,” “South American” or “MesoAmerican.” In order to normalize this, I have combined all of the geographies that are Native indicators.
MDLP
On the MDLP graph below, the legend indicates that these 4 regions are relevant to Native ancestry.
- Army green – Mesoamerican
- Lime Green – Arctic
- Emerald – South American Indian
- Grey – North Amerindian
Chromosome 1 – Native Segment
On the graph below, you can see that mother has more grey than I do from about 162-165, but then I have some grey that she does not at about 170.
A detailed analysis of the segment of chromosome 1 between 158-173 shows the following admixture:
On my results, the putty green, MesoAmerican, is scattered between about 158 and 173, in three segments. The putty green in my mother’s segments are from 159-160.5 and then 167-170.5. Therefore, my father, by inference has a segment from about 162-165 and from about 170.5 to 173.
My teal, North Siberian, ranges from 162-163 and from 168-171. My mother carries no teal in these segments, so this is inferred to be contributed from my father.
My dark grey, North Amerind, ranged from 162-165.5 and then from 168-169.5. My mother’s range is from 161-165.5. Therefore my grey segment at 168-169.5 is either recognized as MesoAmerican or Arctic Amerind in my mother.
Chromosome 2 – Native Segment
Chromosome 2 is quite interesting. You can see that on my chromosome, the North Siberian begins at about 80. Mom has none at that location. My North Amerind begins at about 95 and extends to 105, where Mom’s begins in the same location but then transitions to a large segment of MesoAmerican which I do not carry. I do have MesoAmerican, but mine begins about where hers ends and extends to about 105. Mom’s North Amerind ends about 101, while mine continues to about 105. She looks to have trace amounts beginning about 105 and extending through 115.
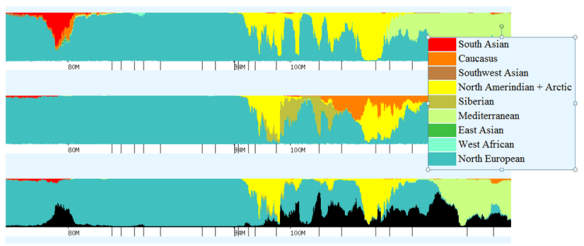
Eurogenes
The next graph shows the same chromosomes using Eurogenes. Regions relevant to Native ancestry include:
- Red – South Asian
- Brown – Southwest Asian
- Yellow – North Amerindian and Arctic
- Putty – Siberian
- Emerald – East Asian
Chromosome 1 – Native Segment
The difference between my chromosome 1 and my mother’s in this region is quite pronounced. My mother’s is drenched in beautiful red South Asian, while I have absolutely none. Some of the area where I have North Amerindian shows as South Asian on hers, but in other areas, there is no correlation. It is expected of course, that there are areas where she has some ancestry and I have none, due to the fact that I only inherit half of her DNA, but she has a significant segment of East Asian between 163 and 164, and I look to have received only a very small portion. The same is true of her Siberian segments at 163-164, but then I have Siberian that she does not at 169-170 and she has some that I don’t at 160-161.5. Some of this difference can likely be explained, especially between the yellow North Amerindian and the red South Asian by slight differences in the DNA read and how it is categorized, but in other cases, the difference is real. Looking at mother’s red segments from about 166.5 to about 168 and then looking at my corresponding region, you can see that I have nothing that hints at Native. In that region, I clearly inherited from my father as well as my mother’s North European.
Chromosome 2 – Native Segment
As different as our chromosomes 1 were, one wouldn’t expect chromosome 2 to be so similar. In the graph, I included my large South Asian segment surrounding 80, where Mom has a trace, although that is beyond the area indicated as Native by 23andMe and Doug McDonald. In the range of interest, beginning at about 80, we find nothing until about 94 where mother and I both have North Amerindian segments that stretch through about 105. Mom’s goes slightly further than mine, to about 105.5. It’s interesting to note that in part of this region, on either side of 101, her Siberian and my North Amerindian are the same shape at the same location, so obviously the same DNA is being read and categorized as two different regions, probably due to my father’s admixture.
Dodecad
On the Dodecad graph of the Native segment, you can see the Native colors are in shades of green.
- Putty – West Asian
- Yellow-green – South Asian
- Emerald – Northeast Asian
- Light Green – Southeast Asian
To use Dodecad in an equivalent manner as the rest of the tools, it looks like Northeast Asian is the closest we would get to Native American since that is where Native Americans lived just prior to crossing Beringia, so the greens should probably be evaluated as a group. As can be seen on chromosome 1, they do clump together. Even though West Asian is also found with this group, it seems to be outside the range, so I am not including it in the evaluation.
Chromosome 1 – Native Segment
You can see another example here of one segment being called South Asian in Mom’s and Northeast Asian in mine at about 170mb.
The Native, or in this case, Northeast Asian/Southeast Asian begins at about 162.5 where Mom’s and mine are very similar. However, we diverge at about 164.5 where Mom begins with large segments of South Asian. I have a little bit, but not much. Beginning about 168, I have a large Northeast Asian segment, but she shows with South Asian there, although the segments are not exact.
Chromosome 2 – Native Segment
Chromsome 2 is quite simple using Dodecad. Only two of the three groups appear. Southeast Asian is absent, South Asian is present only in trace amounts except for one small area between 79.5 and 80 on my chromosome. As expected, Northeast Asia is more prominent. Mother has a few areas that I don’t, which is to be expected.
HarrappaWorld
Last, we have HarrappaWorld. American and Beringian are the Native American categories here. Regions relevant to Native American heritage would be:
- Teal – American
- Periwinkle – Beringian
- Lime Green – Siberia
- Emerald – Northeast Asia
Chromosome 1 – Native Segment
You can see both Beringian and American embedded again at about location 169. In mine, this entire block reads as American.
There is one large chunk of Northeast Asian showing for both results, but part of that region of my chromosome, between 163-164 shows as American instead of Northeast Asian. The Beringian is scattered through the American, which I would expect. The American runs either strongly or weakly through this entire segment from 163 to 175 in mine or to 179 in mother’s. Surprisingly there is no Siberian at all. I would have expected to see Siberian before Northeast Asian.
Chromosome 2 – Native Segment
Where on chromosome 1, we saw no Siberian, on chromosome 2, we find Siberian instead of Northeast Asian. I have no Beringian, but mother has 4 segments. Three of her 4 segments are embedded with American segments. Two may simply be categorized differently in my results, but two, I did not inherit.
Analysis Discussion
What have we learned?
When we are dealing with small amounts of minority admixture, they may or may not be able to be picked up directly by the testing companies. Of course, part of this has to do with their thresholds for what is “real” and reportable, and what isn’t. Aside from that, lack of identification of minority admixture probably has to do with which segments were inherited and their size, if they have been isolated and identified as Native by population geneticists, and the robustness of the data base sources the data is being compared against.
We can also see how difficult it is to sort through threshold matches, meaning what is Native, Asian, central Asian, etc. Many of these differences are probably not actually differences between groups, but similarities with slight categorization differences. Of course, it’s those differences we seek to identify our ancestral heritage. Combining similar geographies may help reveal relationships masked my reporting and categorization differences.
Given that multiple sources have indicated Native ancestry, and on the same two chromosomes, I have no doubt that it exists. Had any doubt remained, the exercises creating the MDLP Chromosome Map Table and reviewing the segments on chromosome 1 between 160 and 180mb would have removed any residual concerns.
The following table shows the results for the Native segments of chromosomes 1 and 2 beginning with the 23andMe and McDonald results, and adding the start and stop segments from each of the 4 admixture tools we used.
| Chromosome 1 | Chromosome 2 | |
| 23andMe |
165,658,091 to 175,711,116 |
86,316,174 to 103,145,426 |
| McDonald |
165,000,000 to 180,000,000 |
90,000,000 to 105,000,000 |
| MDLP |
162,000,000 to 173,000,000 |
80,000,000 to 105,000,000 |
| Eurogenes |
162,500,000 to 171,500,000 |
79,000,000 to 105,000,000 |
| Dodecad? |
162,500,000 to 171,000,000 |
79,500,000 to 105,000,000 |
| Harrappaworld |
163,000,000 to 180,000,000 |
79,000,000 to 104,000,000 |
| In Common |
165,658,091 to 171,000,000 |
90,000,000 to 103,145,426 |
Although the start and end (or stop) segments vary a bit, all resources above confirm that the region on chromosome 1 between 165,658,091 and 171,000,000 is Native and on chromosome 2, between 90,000,000 and 103,145,426. Those are the areas “in common” between all resources, which is shown in the last table entry.
The concept of “in common” is important, because while any one resource may report something differently, or not at all, when all or most of the resources report something the same way, it is less likely to be a fluke or reporting issue, and is much more likely to be real. We’ll be using this methodology throughout the rest of the articles in “The Autosomal Me” series.
In the next segment, Part 8, we’ll be extracting the actual start and stop addresses of the Native only segments, referred to as the “Strong Native” method, and the combined Native indicator segments, referred to as the “Blended Asian” method and looking at how we can use those results.
______________________________________________________________
Disclosure
I receive a small contribution when you click on some of the links to vendors in my articles. This does NOT increase the price you pay but helps me to keep the lights on and this informational blog free for everyone. Please click on the links in the articles or to the vendors below if you are purchasing products or DNA testing.
Thank you so much.
DNA Purchases and Free Transfers
- Family Tree DNA
- MyHeritage DNA only
- MyHeritage DNA plus Health
- MyHeritage FREE DNA file upload
- AncestryDNA
- 23andMe Ancestry
- 23andMe Ancestry Plus Health
- LivingDNA
Genealogy Services
Genealogy Research
- Legacy Tree Genealogists for genealogy research









Thanks for a terrific explanation!
Hi Roberta
I’m following this series with great interest – when it is complete will it be available in toto as a PDF by any chance?
I will probably do that by posting it on my website at http://www.dnaexplain.com under the Publications link.
Pingback: The Autosomal Me – Extracting Data Segments and Clustering | DNAeXplained – Genetic Genealogy
I just downloaded my mother’s results and am trying out the different admixtures. She is definitely showing some Asian and/or Native areas on MDLP and Eurogenes, plus there seems to be some African in quantities that only show up on the chromosome paintings, not on the percentages.
So… here’s the question. We know she’s half Norwegian and a quarter Scottish. The other quarter is unknown (we know her grandfather’s name but nothing else for certain) but I’ve theorized colonial American. Might all the Asian I’m seeing be from the Norwegian (Siberian, etc) or should I boldly assume actual Native American heritage?
For example, Eurogenes K9 gives:
Caucasus4.61%
Southwest Asian1.24%
North Amerindian + Arctic1.13%
Siberian0.18%
Mediterranean21.57%
East Asian0.11%
and the rest is North European.
Yes, Asian can be from an Asiatic source from Europe. I would look at all of the tools and see if you find this consistently. That would be a clue.
Thanks, I’m gradually working my way through. While it would be kind of neat to discover Native American, I don’t want to get ahead of my evidence. If we have actual Native American, it can only be via my one great-grandfather, as everyone else in that generation was fresh off the boat. He would also be the likely bet for the little bits of African that I noticed on MDLP’s chromosome painting.
I meant to ask earlier, how do you handle examining those big long chromosome paintings? I get your basic analytical strategy, it’s the practical side that looks daunting on my screen.
I look at each one the full length and record what I’m looking for as I go. When I was doing this, I could only do it for an hour or so at a time. Be sure to use a large, quality monitor. The thought of trying to do this on a laptop makes me cringe:)
I mainly work on my laptop BUT luckily do have a nice big Dell monitor for visually intensive things. With luck the Dell will also help distinguish between some of the colors that look identical on the laptop.
Even so I’m thinking it would be awfully helpful to be able to compare the various utilities’ chromosome paintings… like get a row of #1 and work through to see how bits of interest are characterized in the different ones.
Plus I’m also wondering at what point one decides something is so little present that it is noise rather than admixture. The African that shows up for my mother is mostly only visible on the chromosome paintings, although some of the charts do give a written percentage. MDLP has some big flares of red (I can’t tell whether it is the Pygmy red or the Sub-Saharan red but it hardly matters which really). As a total beginner at this I’m reading everything I can and trying to figure it out as well as possible!
I listed everything if it was present. If it’s very small, I put “trace” beside it. In some cases, another tool will pick it up well, and you’ll really want to know if it was there or not, even in trace amounts. The first time through I did not do this, but learned when I did the second and third one (for a sanity/insanity check) that I really needed to include trace amounts.
My husband bought me a rather large flat screen TV for my second monitor which I now use constantly.
I really appreciate your thoughts on this. This evening I’ve been reviewing your series (the first time I read it I was fascinated but doubted it would apply to my heritage) and I’m taking an initial look through one of the chromosome 1 images. I think you’re right about making sure to note “trace.” Some of these bits are really tiny, but as you’ve noted, it’s important to check what appears next to what.
This is quite the adventure. We really know nothing about my great-grandfather besides his name; I have a candidate for him but have not been able to tie any autosomal matches to him yet. Until now, everything suggested that he would be all northern European like my recent immigrant ancestors–ie hard to distinguish from anyone else–plus we’ve got no one to do YDNA on. It sure looks to me like there is Native and African in there though. I’m estimating 18th century for that.
I am following along with this series and just recently everything fell in place and the websites working. However, I am already in trouble. How do you suggest starting if you are not a 23&me customer? I have no way to determine what is “real” to start with. FTDNA shows 100% European but most admix utilities at Gedmatch show some Native percentage, especially if Siberian is included. Will McDonald do non-23&me files? Mine is Ancestry, but Mom & Dad are at FTDNA. If he will work with mine, how do I contact him?
I don’t know if Doug accepts Ancestry files or not. Very shortly Family Tree DNA will accept them. Doug’s e-mail is mailto:mcdonald@scs.uiuc.edu
I finally figured out how to tell close colors apart in the admix filters – it is so simple!
If the color flows from the top down it is the one from the top of the color chart. If the color reaches up from the bottom it is the one from the lower portion of the color chart.
Eurogenes K36 in the only one (of those I have looked at) that still has an issue because they repeat some colors 3 times so the mid group still causes problems.
Yeah, I was also baffled with this for a while. Gedmatch should add more colors so the intrepretation would be easier.
Pingback: The Autosomal Me – The Holy Grail – Identifying Native Genealogy Lines | DNAeXplained – Genetic Genealogy
I see quite clearly small amounts of North Amerindian+Artic (Eurogenes K9)in my paintings, on several segments 3-8 and a quite large segment Chr 9 and several of the other paintings as well, but FTdna did not pick it up, I also did ancestrybydna-this is not ancestry.com, anyhow they said I was 19% NA,, with that much you’d think something would show up, but FTdna says 89% Spanish,French,Orcadian, 10.81 Tuscan,Sardinian, and Romanian, very confusing. This is very interesting, I was adopted and only thing I ever heard about birth mother was NA ancestry, so to be able to see that in the dna is important to me, I have found pictures of GG-Grandmother and family stories of NA ancestry, so it was not just a story told to me.
Thank you for helping me to understand this the admixture paintings.
Pingback: The Autosomal Me – Summary and PDF File | DNAeXplained – Genetic Genealogy
Forgive me if this sounds like a stupid question, but is the Gedmatch Chromosome view considered more reliable? When using Admixture Proportions by Chromosome, it shows that I have South, East and West Asian ancestry, as well Native American in rather significant numbers (between 3 and 12% depending upon the Chromosome), when I run it in the Gedmatch Chromosome painting tool, the Asian and Native still show up, but when I run it in Admixture Proportions (with link to Oracle), the Native American percentage, as well as the South and East Asian percentages are lowered below 1% or do not show up at all. Can you shed some light on this? (23andMe didn’t pick up any of the Native or Asian ancestry).
Hi Jay,
I can’t shed light on that, and I would suggest that you contact the creators of the software and ask them.
Roberta
Thanks for responding. I did contact them, but nobody responded to my inquiries.
Jay, if you are on Facebook, there’s a GEDmatch discussion group. Request to join and ask the members there. I can guarantee you will get an answer.
Hello Roberta and fellow-searchers! Love this autosomal DNA series, thank you kindly! I’ve uploaded my sister’s data to GedMatch and have been playing with it (woo-hoo!).
A question: what does it mean to have an ethnicity on more vs. fewer chromosomes, using the “admixture by chromosome” tools? For example, using Eurogenes, we have Amerindian ancestry on 13 chromosomes, ranging from 0.3-4.1%, and we have sub-Saharan African on 5 chromosomes, ranging from 0.3-8.2%. Could this mean the sub-Saharan African genes come in from a smaller branch of the tree? Funny, Ancestry picked up on the African ancestry, but not the Amerindian–that seems to appear as Asian in their breakdown, a common refrain, yeah? Eurogenes also shows a lot of Asian ancestry, particularly West Asian, which is on 18 chromosomes (1.3-21.3%), but Asian and Amerind will be so hard to sort out. We have no known Asian or even Eastern European ancestry, for what it’s worth. All of the Amerindian is on Mom’s side, mostly through her mother’s mother (we have photos of visibly-mixed Civil War era ancestors), but my paternal grandmother was German/Swiss, and ai-ya, you’ve posted about the Germans getting American Indian results. Sad to say my parents have both passed. Here’s hoping when our data is “batched” we’ll find some German cousins to help sort this out! And here’s hoping GedMatch is becoming popular in Germany!
Has anyone found a tribal match with GEDmatch chromosomes charting?